Abstract
Zebrafish have been extensively used for studying vertebrate development and modeling human diseases such as cancer. In the last two decades, they have also emerged as an important model for developmental toxicology research and, more recently, for studying the developmental origins of health and disease (DOHaD). It is widely recognized that epigenetic mechanisms mediate the persistent effects of exposure to chemicals during sensitive windows of development. There is considerable interest in understanding the epigenetic mechanisms associated with DOHaD using zebrafish as a model system. This review summarizes our current knowledge on the effects of environmental chemicals on DNA methylation, histone modifications and noncoding RNAs in the context of DOHaD, and suggest some key considerations in designing experiments for characterizating the mechanisms of action.
Keywords: DNA methylation, noncoding RNAs, DNMTs, zebrafish, non-mammalian model
1. Introduction
There is growing evidence from epidemiological and experimental studies that exposure to environmental stressors during critical windows of susceptibility can have long-term consequences [1,2]. Examples of association between exposure to environmental stressors during critical periods of fetal development and increased risk for cardiovascular diseases, obesity and neurological disorders are well documented [3,4]. This is a growing field of research and is collectively termed as the developmental origins of health and disease (DOHaD) [5,6]. The DOHaD hypothesis postulates that early life stressors can cause developmental reprogramming, inducing long-term changes in normal development and physiology [6]. Several studies have demonstrated that developmental exposure to environmental chemicals can cause long-term changes in physiology and behavior of the adults [7]. Some of these effects are shown to be inherited by subsequent generations. The mechanisms involved in developmental reprogramming by toxicants are not thoroughly understood; however, effects on epigenetic landscape during cellular and tissue differentiation are considered to be a potential mechanism of toxicant action [8]. Epigenetic modifications are defined as persistent changes in gene expression that occur without a change in the nucleotide sequence.
In the past two decades, there has been intense research on the impacts of environmental chemicals on various epigenetic factors using a variety of model systems [9,10]. The majority of the studies were conducted using mammalian models and to a lesser extent in non-mammalian models [11–13]. Agouti mouse is one of the well known model systems used to study epigenetic mechanisms of toxicant action [14]. Even though research in mammalian models can be easily translated to humans, conducting in vivo studies on a rapidly growing list of chemicals is time consuming and not cost-effective. In addition, studying the mechanisms of developmental reprogramming in embryos during in utero development is difficult. Hence, it is increasingly recognized that alternate vertebrate model systems could provide unique advantages in accelerating research in screening toxicants as well as understanding the mechanisms of action. One such model is zebrafish (Danio rerio), an established model in toxicology [15], developmental biology and human disease research [16]. More recently, it has been widely used as a model system for DOHaD studies and for understanding the underlying genetic and epigenetic mechanisms of action [17]. This review summarizes our current knowledge on the epigenetic effects of toxicants using zebrafish as a model organism and highlights the challenges and opportunities zebrafish offers for investigating the epigenetic mechanisms of action. Studies conducted so far have mostly focused on the impact of toxicants on the epigenetic machinery and very little is known about the mechanisms by which toxicants alter the epigenetic patterns. As zebrafish are increasingly used as an alternative model for DOHaD studies, this review summarizes the important factors to consider while conducting studies to characterize the epigenetic basis of DOHaD effects.
Zebrafish as a model for DOHaD and epigenetic toxicology
Zebrafish have become an attractive model for DOHaD and transgenerational studies because of high fecundity, short generation time (embryo to adult in 3–4 months), external fertilization and development, and easy maintenance and breeding [16](Figure 1). In contrast to murine models where embryonic development occurs in utero, in zebrafish it occurs externally. This enables exposure of embryos to stressors immediately after fertilization (2–4 cell stage), in the absence of any maternal influence. Transparent zebrafish embryos allow visualization of any developmental abnormalities associated with exposure. Zebrafish are highly fecund and each female can lay hundreds of eggs at a time. This makes it possible to have relatively high sample size for each experimental condition. There are a number of larval and adult behavioral assays developed to assess the later life effects of developmental exposure to toxicants [18]. Compared to rodent models, rearing and maintenance costs for zebrafish are inexpensive. This is an important consideration for DOHaD and transgenerational studies, because costs associated with raising multiple animals from each treatment condition over a long time period, sometimes over multiple generations, can be expensive. Furthermore, in mammals transgenerational transmission of a phenotype requires assessment of the F3 generation for embryonic exposure because primordial germ cells of the F2 generation are exposed in pregnant dams [19]. In contrast, due to external development in zebrafish, studies in the F2 generation are considered to be transgenerational [17].
Figure 1.
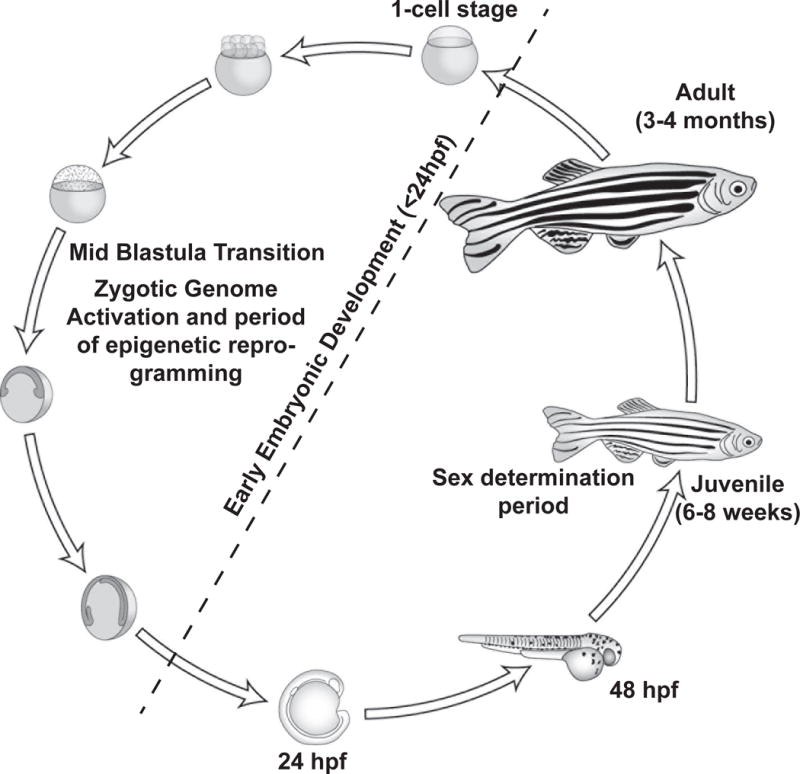
Zebrafish is an ideal model for DOHaD studies. Because of its short life cycle and rapid development they are ideal for conducting long-term studies including multi- and transgenerational studies. Similar to mammals, zebrafish undergo zygotic genome activation as well as epigenetic reprogramming. Exposure to environmental chemicals during sensitive windows of development can have later life consequences.
In addition to these advantages, zebrafish are also ideal for studying the epigenetic mechanisms of action. The availability of numerous transgenic fish strains enables characterization of cell and tissue-specific effects. Zebrafish are also amenable for genetic manipulation, and targeted gene-editing with CRISPR-Cas9 is widely used [20]. The availability of genomic resources [21] and the sequencing methods needed to conduct transcriptomic and epigenomic profiling have garnered enormous attention in the use of zebrafish as a model species in DOHaD studies. In the past few years, there have been several studies characterizing the developmental profiles of DNA methylation [22,23], histone modifications [24] and noncoding RNAs [25–27] providing base line information on the dynamics of epigenetic regulation during embryogenesis. Several studies have demonstrated the long-term effects of developmental exposure to toxicants. Most of these studies have reported later-life effects and in some cases intergenerational or transgenerational effects (Table 1). However, studies aimed at understanding the mechanisms behind DOHaD and multigenerational studies are still in their infancy.
Table 1.
List of published studies investigating the long-term effects of developmental exposure to toxicants in zebrafish.
| Chemical | Exposure | Effects | Reference |
|---|---|---|---|
| TCDD | Developmental and juvenile exposure | Decrease in spermatozoa and germinal epithelium thickness; Increase in spermatogonia |
[66] |
| TCDD | Developmental and juvenile exposure | Increased female to male ratio in all three generations. Scoliosis like phenotype, reduced egg production and fertilization success in F1 and F2 generations. |
[17] |
| TCDD | Developmental exposure | Egg production and fertilization success were reduced Increased mortality of F1 embryos, Reduced egg production and fertility |
[67] |
| Bisphenol A | Adult exposure | Heart defects in F1 and F2 generation | [68] |
| PCB126 | Developmental exposure | Altered adult behavior (lack of habituation) Altered gene expression in the brain |
[69] [70] |
| PAH mixture | Chronic dietary exposure | Altered locomotory activity in F1 and F2 larvae | [71] |
| Testosterone and Dihydrotestosterone | Developmental and juvenile exposure | Global hypomethylation in the ovary (F0) and in F1 larvae. Altered glucose homeostasis |
[72] |
| Atrazine | Developmental exposure | Reduction in 5-hydroxyindoleacetic acid (5-HIAA) levels and serotonin turnover. Reproductive dysfunction in adults |
[71] [73,74] |
| Cadmium | Developmental exposure | Adults displayed anxiety like behavior in novel tank assay Altered antioxidant levels | [75] |
| Vinclozolin | Juvenile exposure | Shift in sex ratios towards females, and affected gonadal maturation. | [76] |
| PFOS | Chronic exposure | Effects on adult behavior (F0) and larval survival, morphology and behavior in F1 generation. | [77] |
| Organophosphate flame retardants | Developmental exposure | Impaired larval and adult behavior | [78] |
Despite the unique advantages zebrafish offer, there are some distinct differences between zebrafish and mammals in epigenetic programming. Mammals undergo two rounds of reprogramming of DNA methylation, first at the time of fertilization in the zygote, and then in primordial germ cells (PGCs). In zebrafish the second wave of reprogramming has not yet been demonstrated. In addition, the methylomes of sperm and oocytes are significantly different and the paternal genome is resistant to demethylation in zebrafish [22,23]. Furthermore, zebrafish do not have genomic imprinting making them unsuitable for studying parent-of-origin effects.
DNA methylation
Similar to mammals, DNA methylation is one of the most well studied epigenetic modifications in zebrafish. Methylation of cytosine residues in CpG islands are generally considered to cause stable silencing of gene expression. Recently, Long et al; [28] empirically demonstrated that CpG islands in gene promoters are conserved among all vertebrates, including zebrafish. DNA methyltransferases (DNMTs) are responsible for the addition of the methyl groups on CpG dinucleotides. Zebrafish possess orthologs of both maintenance and de novo DNMTs [29]. Maintenance DNA methyltransferase, dnmtl, ensures inheritance of methylation patterns during cell division by preferentially methylating hemimethylated CG dinucleotides, whereas de novo methyltransferases are involved in establishing new methylation patterns. In contrast to two de novo DNMTs (DNMT3A and DNMT3B) in mammals, zebrafish possess multiple homologs. These include two DNMT3A (dnmt3aa and dnmt3ab) and four DNMT3B (dnmt3ba, 3bb1, 3bb2 and 3bb3) genes[30]. In general, due to genome duplication in the fish lineage, there are two or more orthologues of many mammalian genes. This often leads to subfunction partitioning among the duplicated genes, providing a unique opportunity for obtaining new mechanistic insights into the multiple functions of a single human gene [31]. For example, DNMT3A knockout mice die postnatally at 4–8 weeks and DNMT3B knockouts die embryonically at 14.5 days making it difficult to study their roles beyond early embryonic development [32]. In contrast, we observed that dnmt3aa and dnmt3ab (DNMT3A homologs) knockout zebrafish generated using TALEN technology develop normally, making it possible to study their roles beyond development. Previous studies using morpholino oligonucleotide knock down approach and TALEN knockouts have demonstrated that proper expression of DNMT3 genes is critical for cellular and tissue differentiation such as hematopoiesis [33]. The results from these studies suggest that toxicants affecting the expression of DNMTs could have long-term consequences by altering DNA methylation. Several studies have investigated the effect of environmental chemicals on DNMT gene expression in zebrafish exposed during early development (Table 2). These studies have shown that altered DNMT gene expression patterns are developmental stage-specific, but whether these changes alter genome wide DNA methylation remains to be determined. So far most of these studies have used gene-specific DNA methylation profiling targeting early developmental genes or those targeting specific pathways and determined alterations to DNA methylation at specific time points [11,34]. Even though these results demonstrate the fact that altered DNMT expression could affect DNA methylation patterns, the prevalence of these changes genomewide are just beginning to be understood. In addition, the persistence of DNA methylation changes observed after developmental exposure remains to be determined. With the availability of high throughput bisulfite sequencing methods, it is feasible to investigate genomewide changes in differential methylation and persistence of these effects over developmental time post-exposure. One recent study demonstrated genomewide transgenerational changes in DNA methylation following developmental exposure to mono (2-ethylhexyl) phthalate in zebrafish [35]. Similar studies needs to be conducted with other toxicants to determine the role of DNA methylation in toxicant-induced phenotypes.
Table 2.
Summary of published studies in zebrafish demonstrating the effects of toxicants on epigenetic mechanisms.
| Chemical | Epigenetic alteration | Developmental stage | References |
|---|---|---|---|
| Estrogen | Aromatase gene promoter DNA methylation | Adults brain and liver | [79] |
| TCDD | DNMT expression Gene-specific DNA methylation | Embryos | [34] |
| BaP | DNMT expression Gene-specific DNA methylation | Embryos | [11,80] |
| Depleted Uranium | Global DNA methylation | Adults | [81,82] |
| TDCPP | Global DNA methylation | Embryos | [83] |
| Lead | Global DNA methylation | Embryos | [84] |
| TCDD | miRA-451, 23a, 23b, 24, 27e | Embryos | [85] |
| Ethanol | miR-9/9*, 153c | Embryos | [86] |
| Ethanol | miR-153a, miR-725, miR-30d, let-7k, miR-100, miR-738, and miR-732 | Embryos | [87] |
| Ethanol | miR-9 | Embryos | [39] |
| Valproic acid | miR-16a, 18c, 122, 132, 457b, and 724 | Embryos | [88] |
Non-coding RNAs
The importance of non-protein coding RNAs (ncRNAs) in the regulation of various biological processes in eukaryotes is well documented. The number of known ncRNAs continues to grow and the most widely known regulatory ncRNAs include microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-associated RNAs (piRNAs) and long noncoding RNAs (lncRNAs). Zebrafish is widely used as a model to study developmental roles of ncRNAs and to determine the mechanisms of regulation of gene expression [26,27,36]. MiRNAs are the most widely studied group in developmental toxicology and a variety of environmental chemicals are shown to affect their expression. However, most of these studies in zebrafish have determined the expression patterns after acute developmental exposure to toxicants; very few studies have characterized the role of individual miRNAs in toxicant-induced phenotypic changes later in life [37–39]. As epigenetic changes are considered to mediate transgenerational effects, it is essential to determine persistent changes that are transmitted to subsequent generations. Recently, a few studies in mice have demonstrated that sperm miRNAs are involved in the transgenerational transmission of stress-induced phenotypes[40,41]. One recent study reported that prenatal exposure of mice to vinclozolin caused an upregulation of miRNA-23b and let-7 in the primordial germ cells of developing embryos [42]. These changes were shown to persist in three subsequent generations. Similar mechanistic studies with other chemicals are needed in order to conclusively demonstrate that miRNAs are a mediator of transgenerational effects. Another group of ncRNAs that are expressed in germ cells and could potentially play a role in transgenerational effects are piRNAs [43]. They originate from the intergenic repetitive elements in the genome and associate with the PIWI subfamily members of the Argonaute family of proteins [43]. Their role is to repress transposable elements in the germline and maintain genomic integrity [44]. The biogenesis of piRNAs in various model systems [45,46], including zebrafish is well known [47–49], but there are no studies on the effects of toxicants on piRNA expression. Recent studies in D. virilis and C. elegans have demonstrated that piRNA-mediated stable long-term gene silencing as a potential mechanism of transgenerational effects [50,51]. However, the hypothesis that ncRNAs act as a mediator of DOHaD and multigenerational effects needs further experimentation.
Histone modifications
Histone modifications play an important role in the regulation of chromatin structure. There is a growing list of these modifications and they exert their effects either by directly influencing the overall chromatin structure or by regulating the binding of effector molecules [52]. For example, methylation of histones can activate (e.g., histone H3 lysine4 trimethylation; H3K4me3) or repress (e.g., histone H3 lysine 27 trimethylation; H3K27me3) gene expression. These histone modifications can alter the chromatin accessibility, thereby generating binding sites for RNA polymerase II or other epigenetic modifiers such as DNMTs and presumably fine-tuning the regulation of gene expression [52]. Similar to the effects on DNA methylation, environmental chemicals that alter the availability of methyl donors can disrupt histone modifications. In addition, histone and DNA demethylases are also shown to impact histone modifications [53]. For example, histone demethylases belonging to Jumonji family are affected by exposure to metals. This is hypothesized to be due to the displacement of iron (Fe), an essential cofactor in the catalytic activity of DNA and histone demethylases by heavy metals [53]. There is some evidence suggesting that altered histone modifications can have persistent effects on gene expression [54,55]. Similar studies are lacking in zebrafish despite considerable progress in characterizing the role of histone modifications during early development and in disease states.
Using genomewide approaches, recent studies have documented the patterns of histone modifications during zygotic genome activation (ZGA), a period of development characterized by major remodeling of chromatin [56]. These results suggest that the ZGA is accompanied by major changes in the patterns of zygotic histone methylation. Based on the evidence that environmental chemical exposures during ZGA affect gene expression patterns, it is conceivable that some of these changes are associated with altered histone modifications. In the past few years there has been significant progress in the development of genomewide profiling methods for analyzing histone modifications in zebrafish [56]. Using these approaches to investigate the effect of environmental chemicals on histone modifications during early embryonic development and their persistence in later life stages is an exciting avenue of research. Furthermore, there is considerable crosstalk between different epigenetic factors. Conducting integrative analysis of gene expression, DNA methylation and histone modifications will be extremely useful to capture the influence of different epigenetic factors on gene expression and the phenotypes.
Considerations for conducting DOHaD studies
Zebrafish have become a popular vertebrate model for studying the long-term implications of exposure to toxicants during sensitive windows of development. Several studies have demonstrated morphological and behavioral phenotypes, but the experiments characterizing the underlying mechanisms are still lacking. Recently, Yamada and Chong [57] described strategies while designing experiments to investigate epigenetic mechanisms of action in DOHaD studies. Some of the factors to consider include the dynamic nature of the epigenome, the relationship between the epigenetic changes and gene expression patterns, cell and tissue-specific differences in the epigenome and the influence of the genome on the epigenetic landscape.
Similar to gene expression patterns, epigenetic changes are dynamic and differ with age. Studies in humans and rodent models have demonstrated that global DNA methylation declines with age [58]. As DOHaD studies investigate the effects a long time after the exposure, it is possible that the environmental factors during the rearing period can influence epigenetic changes. So far most DOHaD studies have investigated epigenetic changes at one time point, often weeks or months after the initial exposure. In order to determine the stable and persistent nature of epigenetic changes, future studies should consider measuring epigenetic changes at multiple time points.
It is also becoming increasingly clear from genomewide profiling studies that the relationship between gene expression and methylation patterns can be ambiguous. Recent studies using genomewide profiling of DNA methylation have shown that the inverse relationship between gene expression and DNA methylation in the promoter regions is not always true [59]. In addition, DNA methylation changes are seen in the intergenic regions far away from any known genes and the functional significance of these changes is yet to be determined.
Most of the DOHaD studies in zebrafish have used gene-specific methods such as quantitative PCR and bisulfite conversion PCR or pyrosequencing for measuring gene expression and DNA methylation, respectively. As the sequencing costs continue to decrease, it is cost-effective to use RNAseq and RRBS (Reduced Representation Bisulfite Sequencing) methods to assess genomewide changes and identify the potential mechanisms. In addition to DNA methylation, other chromatin modifiers such as histones are important players in the regulation of gene expression. Furthermore, the overall chromatin structure is regulated at multiple levels. Determining the nucleosome positioning is a useful approach to initially identify the open and closed chromatin states before conducting targeted analysis of DNA methylation or specific histone modification. Recent development of methods such as Assay for Transposase Accessible Chromatin sequencing (ATAC-seq) have made it less labor intensive to identify regions of open and closed chromatin states, in comparison to Micrococcal Nuclease digestion followed by sequencing (MNase-seq) or DNase hypersensitivity assays [60].
Another important consideration in DOHaD studies is the tissue-specific analysis of epigenetic effects of toxicants. It is well established that despite having the same genome, each cell and tissue type have unique epigenomic patterns. Hence, the effects of toxicant exposure on epigenetic machinery may not be uniform across all tissues and cell types. For example, prenatal bisphenol A exposure in BALB/c mice caused hypermethylation in estrogen receptor 1 gene in the prefrontal cortex, but not in the hypothalamus in male offspring [61]. Therefore, it is important to consider investigating the tissue-specific differences in epigenetic patterns. This may not be possible while working with samples containing heterogenous cell types, such as developing embryos because it is difficult to assign the cellular origin to the sequencing data. However, this problem can be overcome by generating transgenic strains expressing fluorescent proteins in any specific cell type and conducting FACS prior to sequencing. This is particularly feasible in zebrafish where tissue-specific transgenic strains are readily available and methods for FACS are established.
Finally, the most important and poorly understood factor is the role of genetic variation on epigenetic responses. There are widespread associations between single nucleotide polymorphisms (SNPs), the most common source of genetic variation, and DNA methylation in humans [62], but the consequences are poorly understood. It is recently demonstrated that SNPs at CpG sites can impact transcription factor binding [63]. The other well known cause of genomic variation is polymorphisms in the genes involved in the regulation of epigenetic machinery. The effect of genomic variation can be minimized by using inbred strains in DOHaD studies. Even though zebrafish has been used as a laboratory model organism for several decades, the degree of genetic variation among different strains is still quite high [64]. The development of inbred zebrafish lines is necessary in order to reduce confounding effects of genetic variability not only on the epigenome but also on other physiological traits [65].
Conclusions and Future directions
So far most of the DOHaD research using zebrafish as a model system has been focused on identifying the latent effects of developmental exposures. Based on the emerging evidence from mammalian model systems, there is growing recognition that the latent effects of early life exposure could be due to the altered epigenome. In the last few years, there has been a sudden spurt in the number of studies characterizing developmental profiles of various epigenetic factors in zebrafish. These studies suggest that the majority of the epigenetic machinery is highly conserved among vertebrates. In addition, the progress in the development of sequencing techniques and the availability of open source bioinformatic analysis software have made it possible to quantify genomewide epigenetic changes in zebrafish. The next steps in characterizing the epigenetic mechanisms associated with DOHaD include designing experiments that will allow us to identify the developmental basis of persistent changes, and functionally characterizing the genes using forward and/or reverse genetic approaches. Genome editing and transgenesis tools are well established and straightforward, providing unique opportunities to conduct physiological as well as functional studies in an in vivo model system.
Highlights.
Epigenetic mechanisms are highly conserved among all vertebrates.
Zebrafish is an ideal in vivo model for investigating epigenetic mechanisms associated with the developmental origins of adult health and disease.
Among key considerations in conducting DOHaD studies are examination of multiple time-points and tissues, as well as integration of gene expression and methylation analyses.
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences Grant ES024915.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
I wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
I confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed.
I confirm that I have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
I understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. I confirm that we have provided a current, correct email address which is accessible by the Corresponding Author and which has been configured to accept email from
Signed by all authors as follows:
Neel Aluru
naluru@whoi.edu
References
- 1.Barouki R, Gluckman PD, Grandjean P, Hanson M, Heindel JJ. Developmental origins of non-communicable disease: implications for research and public health. Environ Health. 2012;11:42. doi: 10.1186/1476-069X-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heindel JJ, Skalla LA, Joubert BR, Dilworth CH, Gray KA. Review of developmental origins of health and disease publications in environmental epidemiology. Reprod Toxicol. 2017;68:34–48. doi: 10.1016/j.reprotox.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld CS. The Epigenome and Developmental Origins of Health and Disease. Academic Press; 2016. [Google Scholar]
- 4.Schug TT, Barouki R, Gluckman PD, Grandjean P, Hanson M, Heindel JJ. PPTOX III: environmental stressors in the developmental origins of disease–evidence and mechanisms. Toxicol Sci. 2013;131:343–350. doi: 10.1093/toxsci/kfs267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 6.Hanson M. The birth and future health of DOHaD. J Dev Orig Health Dis. 2015;6:434–437. doi: 10.1017/S2040174415001129. [DOI] [PubMed] [Google Scholar]
- 7.Heindel JJ, Balbus J, Birnbaum L, Brune-Drisse MN, Grandjean P, Gray K, Landrigan PJ, Sly PD, Suk W, Cory Slechta D, Thompson C, et al. Developmental Origins of Health and Disease: Integrating Environmental Influences. Endocrinology. 2015;156:3416–3421. doi: 10.1210/EN.2015-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skinner MK. Endocrine disruptors in 2015: Epigenetic transgenerational inheritance. Nat Rev Endocrinol. 2016;12:68–70. doi: 10.1038/nrendo.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaze J, Roth TL. Evidence from clinical and animal model studies of the long-term and transgenerational impact of stress on DNA methylation. Semin Cell Dev Biol. 2015;43:76–84. doi: 10.1016/j.semcdb.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenfeld CS. Animal models to study environmental epigenetics. Biol Reprod. 2010;82:473–488. doi: 10.1095/biolreprod.109.080952. [DOI] [PubMed] [Google Scholar]
- 11.Corrales J, Fang X, Thornton C, Mei W, Barbazuk WB, Duke M, Scheffler BE, Willett KL. Effects on specific promoter DNA methylation in zebrafish embryos and larvae following benzo[a]pyrene exposure. Comp Biochem Physiol C Toxicol Pharmacol. 2014;163:37–46. doi: 10.1016/j.cbpc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asselman J, De Coninck DI, Beert E, Janssen CR, Orsini L, Pfrender ME, Decaestecker E, De Schamphelaere KA. Bisulfite Sequencing with Daphnia Highlights a Role for Epigenetics in Regulating Stress Response to Microcystis through Preferential Differential Methylation of Serine and Threonine Amino Acids. Environ Sci Technol. 2017;51:924931. doi: 10.1021/acs.est.6b03870. [DOI] [PubMed] [Google Scholar]
- 13.Tejeda-Benitez L, Olivero-Verbel J. Caenorhabditis elegans, a Biological Model for Research in Toxicology. Rev Environ Contam Toxicol. 2016;237:1–35. doi: 10.1007/978-3-319-23573-8_1. [DOI] [PubMed] [Google Scholar]
- 14.Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 15.Garcia GR, Noyes PD, Tanguay RL. Advancements in zebrafish applications for 21st century toxicology. Pharmacol Ther. 2016;161:11–21. doi: 10.1016/j.pharmthera.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunwald DJ, Eisen JS. Headwaters of the zebrafish – emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 17.Baker TR, Peterson RE, Heideman W. Using zebrafish as a model system for studying the transgenerational effects of dioxin. Toxicol Sci. 2014;138:403–411. doi: 10.1093/toxsci/kfu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart AM, Braubach O, Spitsbergen J, Gerlai R, Kalueff AV. Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 2014;37:264–278. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackburn PR, Campbell JM, Clark KJ, Ekker SC. The CRISPR system–keeping zebrafish gene targeting fresh. Zebrafish. 2013;10:116–118. doi: 10.1089/zeb.2013.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, Giron CG, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, Zhang J, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153:759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrup O, Reiner AH, Alestrom P, Collas P. The specific alteration of histone methylation profiles by DZNep during early zebrafish development. Biochim Biophys Acta. 2014;1839:1307–1315. doi: 10.1016/j.bbagrm.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 26.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 27.Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grutzner F, Odom DT, Patient R, Ponting CP, Klose RJ. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. 2013;2:e00348. doi: 10.7554/eLife.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goll MG, Halpern ME. DNA methylation in zebrafish. Prog Mol Biol Transl Sci. 2011;101:193–218. doi: 10.1016/B978-0-12-387685-0.00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campos C, Valente LM, Fernandes JM. Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene. 2012;500:93–100. doi: 10.1016/j.gene.2012.03.041. [DOI] [PubMed] [Google Scholar]
- 31.Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 33.Gore AV, Athans B, Iben JR, Johnson K, Russanova V, Castranova D, Pham VN, Butler MG, Williams-Simons L, Nichols JT, Bresciani E, et al. Epigenetic regulation of hematopoiesis by DNA methylation. Elife. 2016;5:e11813. doi: 10.7554/eLife.11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aluru N, Kuo E, Helfrich LW, Karchner SI, Linney EA, Pais JE, Franks DG. Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio) Toxicol Appl Pharmacol. 2015;284:142–151. doi: 10.1016/j.taap.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamstra JH, Sales LB, Alestrom P, Legler J. Differential DNA methylation at conserved non-genic elements and evidence for transgenerational inheritance following developmental exposure to mono(2-ethylhexyl) phthalate and 5-azacytidine in zebrafish. Epigenetics Chromatin. 2017;10:20. doi: 10.1186/s13072-017-0126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schier AF, Giraldez AJ. MicroRNA function and mechanism: insights from zebra fish. Cold Spring Harb Symp Quant Biol. 2006;71:195–203. doi: 10.1101/sqb.2006.71.055. [DOI] [PubMed] [Google Scholar]
- 37.Franzosa JA, Bugel SM, Tal TL, La Du JK, Tilton SC, Waters KM, Tanguay RL. Retinoic acid-dependent regulation of miR-19 expression elicits vertebrate axis defects. FASEB J. 2013;27:4866–4876. doi: 10.1096/fj.12-225524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khayrullin A, Smith L, Mistry D, Dukes A, Pan YA, Hamrick MW. Chronic alcohol exposure induces muscle atrophy (myopathy) in zebrafish and alters the expression of microRNAs targeting the Notch pathway in skeletal muscle. Biochem Biophys Res Commun. 2016;479:590–595. doi: 10.1016/j.bbrc.2016.09.117. [DOI] [PubMed] [Google Scholar]
- 39.Pappalardo-Carter DL, Balaraman S, Sathyan P, Carter ES, Chen WJ, Miranda RC. Suppression and epigenetic regulation of MiR-9 contributes to ethanol teratology: evidence from zebrafish and murine fetal neural stem cell models. Alcohol Clin Exp Res. 2013;37:1657–1667. doi: 10.1111/acer.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, Donkin I, Sjogren R, Mudry JM, Vetterli L, Gupta S, et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab. 2016;5:184–197. doi: 10.1016/j.molmet.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodgers AB, Morgan CP, Leu NA, Bale TL. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A. 2015;112:13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brieno-Enriquez MA, Garcia-Lopez J, Cardenas DB, Guibert S, Cleroux E, Ded L, Hourcade Jde D, Peknicova J, Weber M, Del Mazo J. Exposure to endocrine disruptor induces transgenerational epigenetic deregulation of microRNAs in primordial germ cells. PLoS One. 2015;10:e0124296. doi: 10.1371/journal.pone.0124296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weick EM, Miska EA. piRNAs: from biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 44.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:23612373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Albuquerque BF, Placentino M, Ketting RF. Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev Cell. 2015;34:448–456. doi: 10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Rosenkranz D, Han CT, Roovers EF, Zischler H, Ketting RF. Piwi proteins and piRNAs in mammalian oocytes and early embryos: From sample to sequence. Genom Data. 2015;5:309–313. doi: 10.1016/j.gdata.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang HY, Houwing S, Kaaij LJ, Meppelink A, Redl S, Gauci S, Vos H, Draper BW, Moens CB, Burgering BM, Ladurner P, et al. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 2011;30:3298–3308. doi: 10.1038/emboj.2011.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29:3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erwin AA, Galdos MA, Wickersheim ML, Harrison CC, Marr KD, Colicchio JM, Blumenstiel JP. piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of D. virilis. PLoS Genet. 2015;11:e1005332. doi: 10.1371/journal.pgen.1005332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minkina O, Hunter CP. Stable Heritable Germline Silencing Directs Somatic Silencing at an Endogenous Locus. Mol Cell. 2017;65:659–670 e655. doi: 10.1016/j.molcel.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chervona Y, Arita A, Costa M. Carcinogenic metals and the epigenome: understanding the effect of nickel, arsenic, and chromium. Metallomics. 2012;4:619–627. doi: 10.1039/c2mt20033c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jefferson WN, Chevalier DM, Phelps JY, Cantor AM, Padilla-Banks E, Newbold RR, Archer TK, Kinyamu HK, Williams CJ. Persistently altered epigenetic marks in the mouse uterus after neonatal estrogen exposure. Mol Endocrinol. 2013;27:1666–1677. doi: 10.1210/me.2013-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q, Trevino LS, Wong RL, Medvedovic M, Chen J, Ho SM, Shen J, Foulds CE, Coarfa C, O’Malley BW, Shilatifard A, et al. Reprogramming of the Epigenome by MLL1 Links Early-Life Environmental Exposures to Prostate Cancer Risk. Mol Endocrinol. 2016;30:856871. doi: 10.1210/me.2015-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Gomez-Skarmeta JL. The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods. 2013;62:207–215. doi: 10.1016/j.ymeth.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Yamada L, Chong S. Epigenetic studies in Developmental Origins of Health and Disease: pitfalls and key considerations for study design and interpretation. J Dev Orig Health Dis. 2017;8:30–43. doi: 10.1017/S2040174416000507. [DOI] [PubMed] [Google Scholar]
- 58.Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol. 2015;13:7. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chernyavskaya Y, Kent B, Sadler KC. Zebrafish Discoveries in Cancer Epigenetics. Adv Exp Med Biol. 2016;916:169–197. doi: 10.1007/978-3-319-30654-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kundakovic M, Gudsnuk K, Franks B, Madrid J, Miller RL, Perera FP, Champagne FA. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc Natl Acad Sci U S A. 2013;110:9956–9961. doi: 10.1073/pnas.1214056110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Volkov P, Olsson AH, Gillberg L, Jorgensen SW, Brons C, Eriksson KF, Groop L, Jansson PA, Nilsson E, Ronn T, Vaag A, et al. A Genome-Wide mQTL Analysis in Human Adipose Tissue Identifies Genetic Variants Associated with DNA Methylation, Gene Expression and Metabolic Traits. PLoS One. 2016;11:e0157776. doi: 10.1371/journal.pone.0157776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banovich NE, Lan X, McVicker G, van de Geijn B, Degner JF, Blischak JD, Roux J, Pritchard JK, Gilad Y. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10:e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guryev V, Koudijs MJ, Berezikov E, Johnson SL, Plasterk RH, van Eeden FJ, Cuppen E. Genetic variation in the zebrafish. Genome Res. 2006;16:491–497. doi: 10.1101/gr.4791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butler MG, Iben JR, Marsden KC, Epstein JA, Granato M, Weinstein BM. SNPfisher: tools for probing genetic variation in laboratory-reared zebrafish. Development. 2015;142:1542–1552. doi: 10.1242/dev.118786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baker BB, Yee JS, Meyer DN, Yang D, Baker TR. Histological and Transcriptomic Changes in Male Zebrafish Testes Due to Early Life Exposure to Low Level 2,3,7,8-Tetrachlorodibenzo-p-Dioxin. Zebrafish. 2016;13:413–423. doi: 10.1089/zeb.2016.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.King Heiden TC, Spitsbergen J, Heideman W, Peterson RE. Persistent adverse effects on health and reproduction caused by exposure of zebrafish to 2,3,7,8-tetrachlorodibenzo-p-dioxin during early development and gonad differentiation. Toxicol Sci. 2009;109:7587. doi: 10.1093/toxsci/kfp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lombo M, Fernandez-Diez C, Gonzalez-Rojo S, Navarro C, Robles V, Herraez MP. Transgenerational inheritance of heart disorders caused by paternal bisphenol A exposure. Environ Pollut. 2015;206:667–678. doi: 10.1016/j.envpol.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 69.Glazer L, Hahn ME, Aluru N. Delayed effects of developmental exposure to low levels of the aryl hydrocarbon receptor agonist 3,3′,4,4′,5-pentachlorobiphenyl (PCB126) on adult zebrafish behavior. Neurotoxicology. 2016;52:134–143. doi: 10.1016/j.neuro.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aluru N, Glazer L, Hahn ME. Early-Life Exposure to Low Levels of AHR Agonist 3,3′,4,4′,5-Pentachlorobiphenyl (PCB126) Reprograms Gene Expression in Adult Brain. The Toxicologist: Supplement to Toxicological Sciences. 2017:156. doi: 10.1093/toxsci/kfx192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vignet C, Joassard L, Lyphout L, Guionnet T, Goubeau M, Le Menach K, Brion F, Kah O, Chung BC, Budzinski H, Begout ML, et al. Exposures of zebrafish through diet to three environmentally relevant mixtures of PAHs produce behavioral disruptions in unexposed F1 and F2 descendant. Environ Sci Pollut Res Int. 2015;22:16371–16383. doi: 10.1007/s11356-015-4157-8. [DOI] [PubMed] [Google Scholar]
- 72.Xu N, Chua AK, Jiang H, Liu NA, Goodarzi MO. Early embryonic androgen exposure induces transgenerational epigenetic and metabolic changes. Mol Endocrinol. 2014;28:1329–1336. doi: 10.1210/me.2014-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wirbisky SE, Sepulveda MS, Weber GJ, Jannasch AS, Horzmann KA, Freeman JL. Embryonic Atrazine Exposure Elicits Alterations in Genes Associated with Neuroendocrine Function in Adult Male Zebrafish. Toxicol Sci. 2016;153:149–164. doi: 10.1093/toxsci/kfw115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wirbisky SE, Weber GJ, Sepulveda MS, Lin TL, Jannasch AS, Freeman JL. An embryonic atrazine exposure results in reproductive dysfunction in adult zebrafish and morphological alterations in their offspring. Sci Rep. 2016;6:21337. doi: 10.1038/srep21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruiter S, Sippel J, Bouwmeester MC, Lommelaars T, Beekhof P, Hodemaekers HM, Bakker F, van den Brandhof EJ, Pennings JL, van der Ven LT. Programmed Effects in Neurobehavior and Antioxidative Physiology in Zebrafish Embryonically Exposed to Cadmium: Observations and Hypothesized Adverse Outcome Pathway Framework. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17111830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lor Y, Revak A, Weigand J, Hicks E, Howard DR, King-Heiden TC. Juvenile exposure to vinclozolin shifts sex ratios and impairs reproductive capacity of zebrafish. Reprod Toxicol. 2015;58:111–118. doi: 10.1016/j.reprotox.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 77.Chen J, Das SR, La Du J, Corvi MM, Bai C, Chen Y, Liu X, Zhu G, Tanguay RL, Dong Q, Huang C. Chronic PFOS exposures induce life stage-specific behavioral deficits in adult zebrafish and produce malformation and behavioral deficits in F1 offspring. Environ Toxicol Chem. 2013;32:201–206. doi: 10.1002/etc.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliveri AN, Bailey JM, Levin ED. Developmental exposure to organophosphate flame retardants causes behavioral effects in larval and adult zebrafish. Neurotoxicol Teratol. 2015;52:220–227. doi: 10.1016/j.ntt.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stromqvist M, Tooke N, Brunstrom B. DNA methylation levels in the 5′ flanking region of the vitellogenin I gene in liver and brain of adult zebrafish (Danio rerio)–sex and tissue differences and effects of 17alpha-ethinylestradiol exposure. Aquat Toxicol. 2010;98:275–281. doi: 10.1016/j.aquatox.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 80.Fang X, Corrales J, Thornton C, Scheffler BE, Willett KL. Global and gene specific DNA methylation changes during zebrafish development. Comp Biochem Physiol B Biochem Mol Biol. 2013;166:99–108. doi: 10.1016/j.cbpb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gombeau K, Bourdineaud JP, Ravanat JL, Armant O, Camilleri V, Cavalie I, Floriani M, Adam-Guillermin C. Epigenetic, histopathological and transcriptomic effects following exposure to depleted uranium in adult zebrafish and their progeny. Aquat Toxicol. 2017;184:14–25. doi: 10.1016/j.aquatox.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 82.Gombeau K, Pereira S, Ravanat JL, Camilleri V, Cavalie I, Bourdineaud JP, Adam-Guillermin C. Depleted uranium induces sex- and tissue-specific methylation patterns in adult zebrafish. J Environ Radioact. 2016;154:25–33. doi: 10.1016/j.jenvrad.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 83.McGee SP, Cooper EM, Stapleton HM, Volz DC. Early zebrafish embryogenesis is susceptible to developmental TDCPP exposure. Environ Health Perspect. 2012;120:1585–1591. doi: 10.1289/ehp.1205316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanchez OF, Lee J, Yu King Hing N, Kim SE, Freeman JL, Yuan C. Lead (Pb) exposure reduces global DNA methylation level by non-competitive inhibition and alteration of dnmt expression. Metallomics. 2017;9:149–160. doi: 10.1039/c6mt00198j. [DOI] [PubMed] [Google Scholar]
- 85.Jenny MJ, Aluru N, Hahn ME. Effects of short-term exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin on microRNA expression in zebrafish embryos. Toxicol Appl Pharmacol. 2012;264:262–273. doi: 10.1016/j.taap.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tal TL, Franzosa JA, Tilton SC, Philbrick KA, Iwaniec UT, Turner RT, Waters KM, Tanguay RL. MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J. 2012;26:1452–1461. doi: 10.1096/fj.11-194464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soares AR, Pereira PM, Ferreira V, Reverendo M, Simoes J, Bezerra AR, Moura GR, Santos MA. Ethanol exposure induces upregulation of specific microRNAs in zebrafish embryos. Toxicol Sci. 2012;127:18–28. doi: 10.1093/toxsci/kfs068. [DOI] [PubMed] [Google Scholar]
- 88.Aluru N, Deak KL, Jenny MJ, Hahn ME. Developmental exposure to valproic acid alters the expression of microRNAs involved in neurodevelopment in zebrafish. Neurotoxicol Teratol. 2013;40:46–58. doi: 10.1016/j.ntt.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


