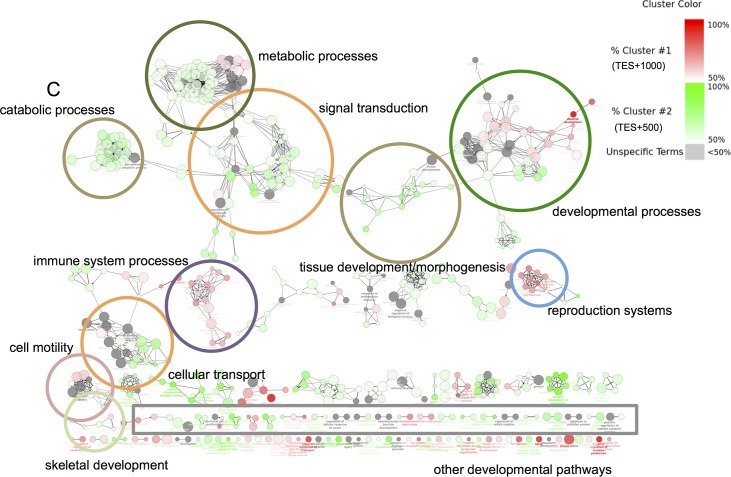
Fig 7. Genes with differentially hypersensitive 3’ regions near TES+1000 or TES+500 strongly correlate with developmental gene ontology pathways.
Ontology term enrichment analysis of GO_Biological Processes was performed on the group of genes clustered by the presence of DHS peaks centered near TES+1000 or TES+500 sequences. Each node represents an enriched ontology term. Enrichment terms that were defined by 51% or more by TES+1000 genes are shaded in red, while terms defined by 51% or more by the TES+500 cluster of genes are shaded in green. Color intensities of each node reflect the relative strength they are enriched by either group. Gray nodes are terms equally enriched by both gene groups (Unspecified terms). The gray box represents ontology terms associated with various developmental pathways. Term nodes related to developmental processes, reproductive systems, and immune response processes are defined more by TES+1000 genes, while term nodes for metabolic and catabolic processes, signal transduction, cellular transport, and tissue or morphogenesis processes are defined more by the TES+500 genes.