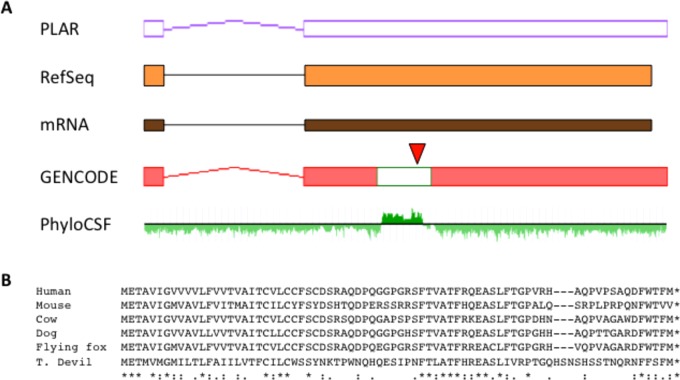
Figure 4.
Reannotation of OTTHUMG00000019887 based on proteogenomic analysis. (A) This locus was present in GENCODE v20 as a lincRNA model, and it is currently categorized in this way by RefSeq (orange model) based on mRNA AK056723.1 (brown model) and given the official HGNC gene symbol LINC00961. Furthermore, an equivalent model was generated and classified as a lncRNA by the RNA-Seq-based PLAR pipeline developed by Hezroni et al.38 (purple-outlined model). GENCODE have now converted this model to protein coding (UTRs in red; CDS in green) based on proteogenomic evidence in combination with evolutionary conservation. The conserved region is well resolved by PhyloCSF, with this track being taken from genome.ucsc.edu. Peptide [QEASLFTGPVR] is marked (red triangle). (B) The 75 aa human CDS shows conservation in eutherian mammals, although not outside this group based on available genome alignments. “T. Devil” is Tasmanian devil, and “flying fox” is specifically the black flying fox Pteropus alecto.