Figure 2.
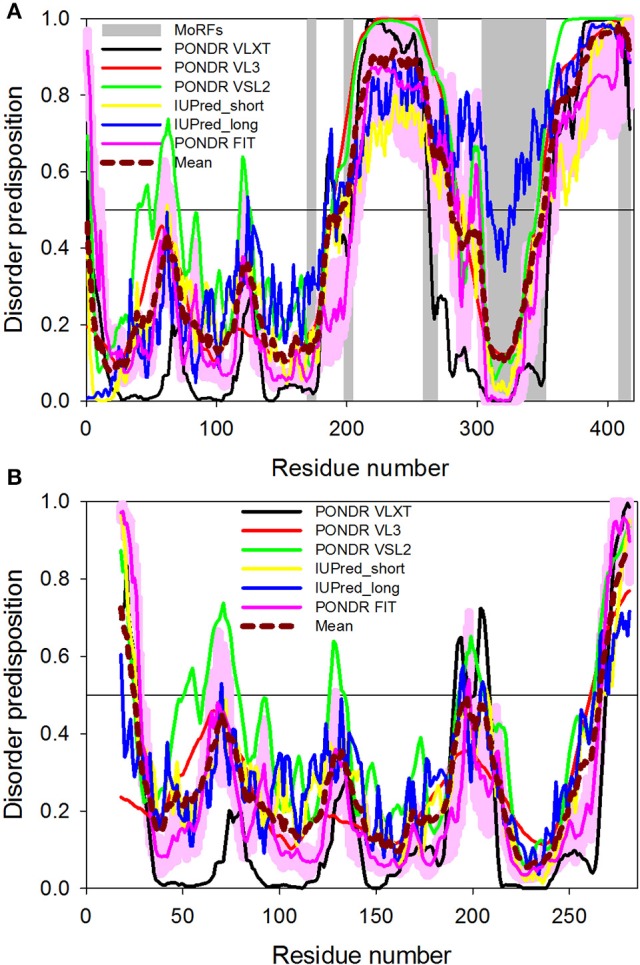
Intrinsic disorder propensity of wild-type human CALR (A) and of the construct of human CALR used in the crystallization study. (A) Evaluating intrinsic disorder propensity of human CALR (UniProt ID: P27797) by series of per-residue disorder predictors. Disorder profiles generated by PONDR® VLXT, PONDR FIT, PONDR® VL3, PONDR® VSL2, IUPred_short, and IUPred_long are shown by black, red, green, yellow, blue, and pink lines, respectively. Dashed dark red shows the mean disorder propensity calculated by averaging disorder profiles of individual predictors. Light pink shadow around the PONDR® FIT shows error distribution. In these analyses, the predicted intrinsic disorder scores above 0.5 are considered to correspond to the disordered residues/regions. Positions of the MoRF regions are shown as gray shaded areas. Modified from Migliaccio and Uversky (2017). (B) Intrinsic disorder propensity of the crystallized construct of human CALR described in Figure 3A by series of per-residue disorder predictors. The amino acid sequence of this construct includes the N-domain (residues 18–204) and the N-terminal half of the C-domain (residues 303–368) connected by a GSG tripeptide. Disorder profiles generated by the same program described in (A) [see legend of (A) for further details]. See Migliaccio and Uversky (2017).
