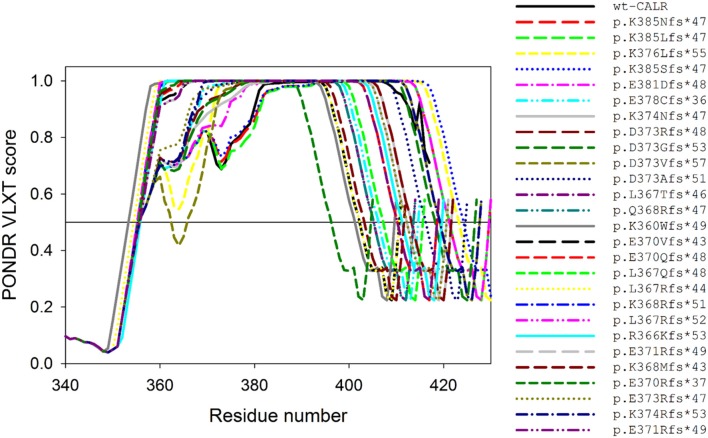
Figure 4.
Effects of the indel mutations found in myeloproliferative disorders on the intrinsic disorder predisposition of human CALR. Shown are disorder profiles generated for wild-type CALR and its indel mutations by PONDR® VLXT algorithm (Romero et al., 2001). Since these mutations do not have any effect on the N-CALR, P-domain, and the N-terminal half of C-CALR domain, the plot zooms in the C-terminal part of the C-CALR domain. We used the PONDR® VLXT program because, although is not the most accurate disorder predictor, is sensitive to local peculiarities of the AA sequence and therefore is the program of choice to identify disorder-based potential functional sites. The multiparametic analysis of intrinsic disorder predisposition of wild-type CALR is shown also shown in Figure 2 and is published in Migliaccio and Uversky (2017). See Migliaccio and Uversky (2017).