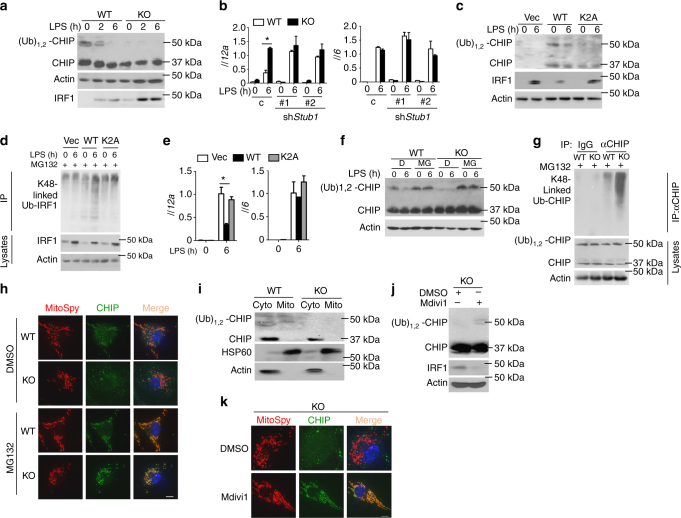
Fig. 6.
CHIP monoubiquitination is involved in IRF1 degradation. a IB analysis of CHIP and monoubiquitinated CHIP in the cytoplasm and IRF1 in the nucleus after stimulation with LPS as indicated. b WT or FAM73b KO BMDMs were infected with shRNA (C) or two different Stub1-specific shRNAs. qRT-PCR analysis of the indicated cytokines. c, d CHIP-deficient RAW264.7 cells were reconstituted with WT and mutant CHIP. c IB analyses of the indicated proteins in subcellular fractions. d IRF1 polyubiquitination in whole-cell lysates. e qRT-PCR analysis of Il12a and Il6 mRNA in reconstituted RAW264.7 cells. f, g WT and FAM73b KO BMDMs were treated with MG132 for 30 min before being collected. IB analyses of CHIP monoubiquitination in the cytoplasm (f) and the polyubiquitination level (g). h Confocal microscopy analyses of CHIP location. i Immunoblot analysis of CHIP in mitochondrial or non-mitochondrial extracts. The purity of the mitochondrial fraction was confirmed using antibodies targeting HSP60 (a mitochondria marker) and Actin. j, k FAM73b KO BMDMs were treated with Mdivi-1 for 12 h. The protein level and location of CHIP were measured by IB (j) and confocal microscopy (k). The data are representative of three independent experiments. Scale bars in h and k are 10 μm. Statistical analyses represent variations in experimental replicates. Two-tailed unpaired t-tests were performed; *P < 0.05