FIG 2.
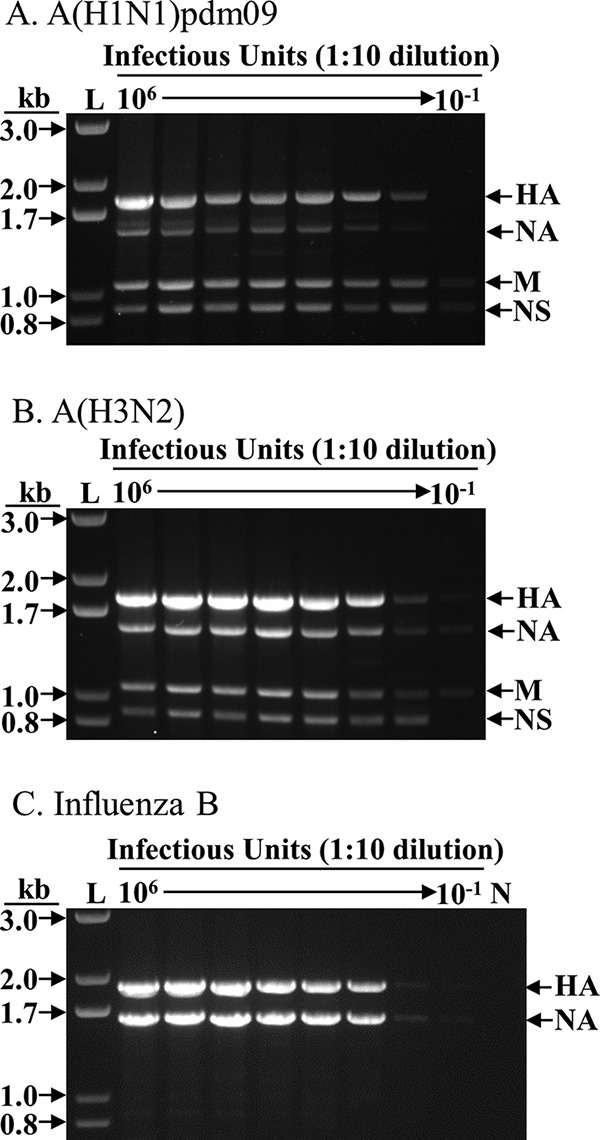
Sensitivity of FluA/B surveillance multiplex RT-PCR. RNA was extracted from 107 TCID50 of each A(H1N1)pdm09, A(H3N2), and influenza B virus, eluted in 30 μl of nuclease-free water, and diluted in a 1:10 series; 3 μl was used as the template for each RT-PCR. The equivalent amount of virus (TCID50) used in each RT-PCR is shown at the top of each lane. For each 25-μl reaction mixture, 3 μl was used for electrophoresis in agarose gel. L, 1-kb Plus ladder (Thermo Fisher Scientific, Inc.); N, negative control (no template). (A) A/New York/NY1682/2009 (H1N1pdm09). (B) A/New York/238/2005 (H3N2). (C) B/Brisbane/60-10/2010 (influenza B virus). The particular A(H1N1)pdm09 virus used here was rescued in an earlier study (13) and contains a “G” at the fourth position of the NA segment (refer to Fig. 1), which may have resulted in the less-robust amplification of the NA segment, a phenomenon we have not noticed in the many wild-type A(H1N1)pdm09 viruses we processed (Table 2).
