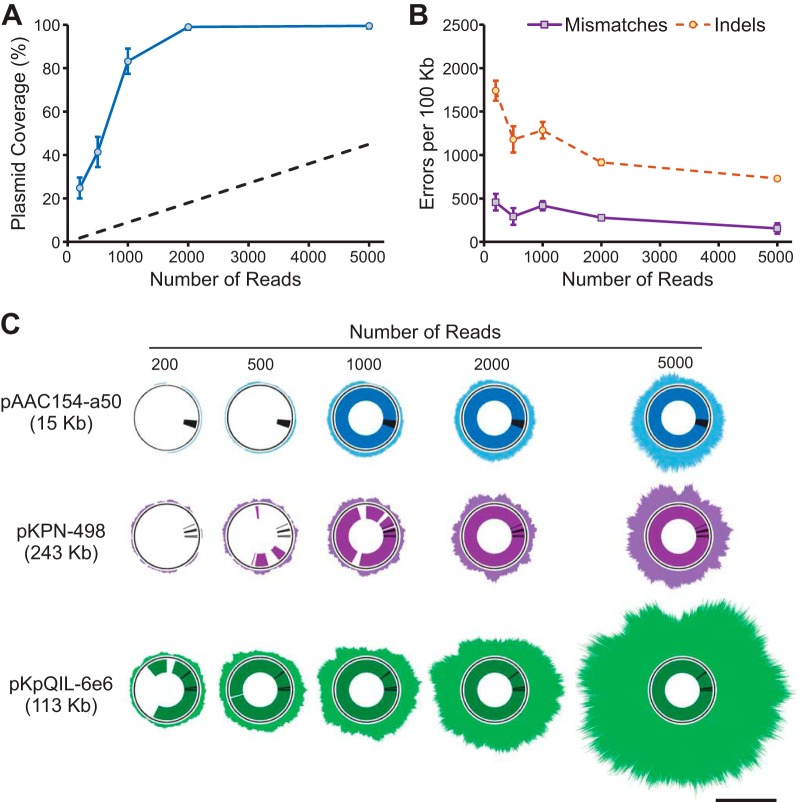
FIG 1.
Reference genome coverage and resistance gene detection in data subsets of KPNIH1. Random subsets of MinION reads (200, 500, 1,000, 2,000, and 5,000 reads) were selected to evaluate minimum requirements for plasmid assembly and resistance gene prediction. (A) Overall KPNIH1 plasmid coverage by assembled contigs (means ± SDs calculated over 10 random samples) at each sampled read count. Dashed lines indicates rough estimate of plasmid coverage achieved by sequencing of total DNA, calculated as straight-line projection from measured plasmid coverage data points and assuming ∼10% plasmid content. (B) Assembly accuracy (means ± SDs calculated over 10 random samples) for each data subset. (C) Reference genome coverage by raw MinION reads and assembled contigs for reach data subset. Outer rings represent raw read alignment to reference plasmid assemblies; inner rings represent percent reference sequence covered by assembled contigs. Black lines in inner rings indicate positions of resistance genes for reference. The scale bar indicates 100-fold sequencing depth.