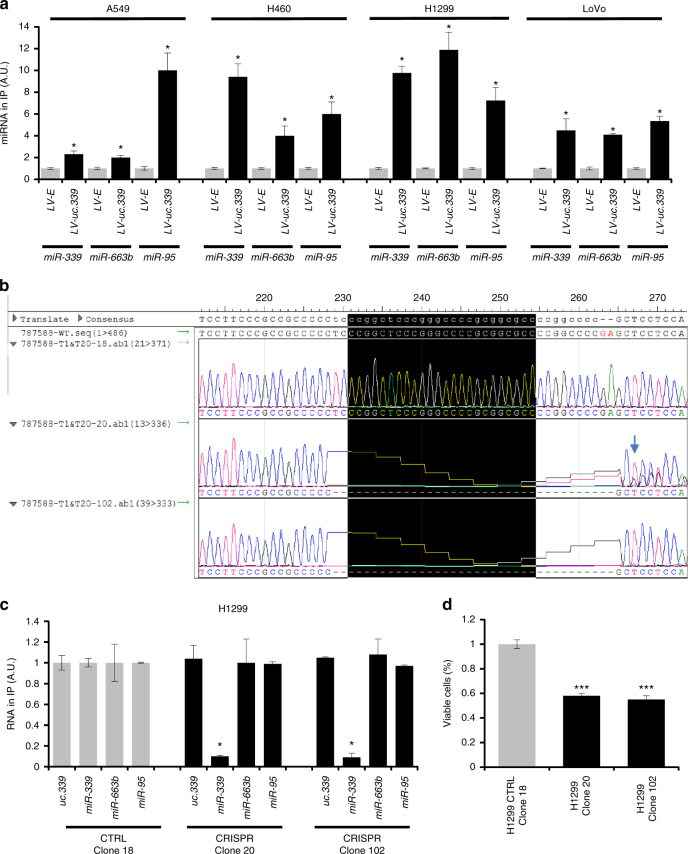
Fig. 6.
Endogenous uc.339 sequesters miRNAs. a qRT-PCR for miR-339, -663b, and -95 in A549, H460, H1299, and LoVo cells infected with LV-uc.339 or LV-E, in which uc.339 has been immunoprecipitated. The expression of miRNAs is presented as normalized to the LV-E group. Data are shown as mean ± s.d. of experiments conducted in triplicate. *P < 0.05. b Sequencing of clones #18, #20 and #102 in H1299 cells engineered with CRISPR/Cas9 genomic technology for the deletion of the binding site of miR-339 (nucleotides inside the black box) in the sequence of uc.339. c qRT-PCR for uc.339, miR-339, -663b, and -95 in the immunoprecipitate of uc.339 in H1299 cells wild-type (clone 18) or in clones #20 and #102 of H1299 in which the MBE for miR-339 has been deleted with CRISPR/Cas9 technology. The expression of the RNAs is presented as normalized to the clone 18 group. Data are shown as mean ± s.d. of experiments conducted in triplicate. *P < 0.05. d Cell viability assay in H1299 cells expressing wild-type uc.339 (H1299 CTRL Clone 18) or in CRISPR clones 20 and 102 after 72 h in culture. Data are presented as mean ± s.d. of experiments conducted in triplicate and normalized to H1299 wt. *P < 0.05