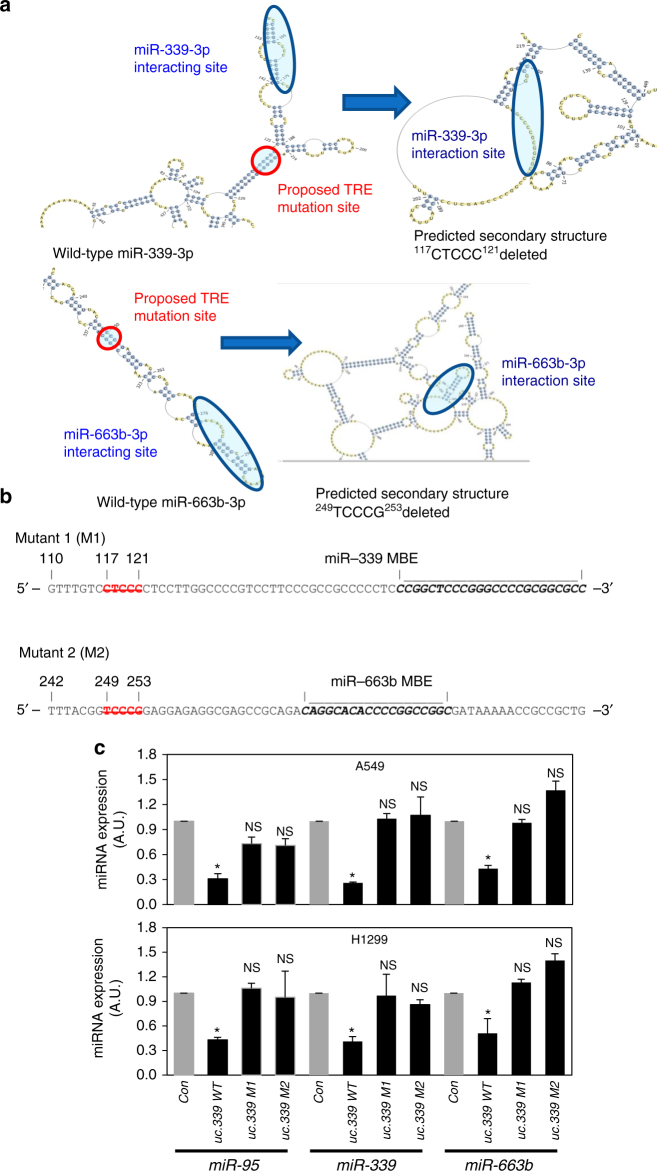
Fig. 8.
Identification of trapping-related elements (TRE) in the uc.339 transcript. a Proposed local structural effect of deletions in TRE in the uc.339 RNA transcript by using RNAfold from the ViennaRNA Package. b Mutants of the TRE sequences in the uc.339 transcript. The TREs are indicated in red, and their position related to the beginning of the uc.339 transcript (as in Supplementary Fig. 1c) is indicated by the numbers above. In bold, the predicted miRNA-binding elements (MBEs) for miR-339 or miR-663b. The figure shows which specific bases were deleted to generate mutant 1 (M1) and mutant 2 (M2) for the experiments in Fig. 8c. c qRT-PCR for miR-95, -339, and -663b in A549 and H1299 cells untreated (Con) or transfected with a plasmid-expressing wild-type uc.339 (uc.339 WT) or TRE M1-mutated uc.339 (uc.339 M1) or TRE M2-mutated uc.339 (uc.339 M2). The expression of miRNAs is presented as normalized to the Con group. Data are shown as mean ± s.d. of experiments conducted in triplicate. *P < 0.05. NS not significant