Abstract
Pseudotrichonympha is a large and structurally complex genus of parabasalian protists that play a key role in the digestion of lignocellulose in the termite hindgut. Like many termite symbionts, it has a conspicuous body plan that makes genus-level identification relatively easy, but species-level diversity of Pseudotrichonympha is understudied. Molecular surveys have suggested the diversity is much greater than the current number of described species, and that many “species” described in multiple hosts are in fact different, but gene sequences from formally described species remain a rarity. Here we describe three new species from Coptotermes and Prorhinotermes hosts, including small subunit ribosomal RNA (SSU rRNA) sequences from single cells. Based on host identification by morphology and DNA barcoding, as well as the morphology and phylogenetic position of each symbiont, all three represent new Pseudotrichonympha species: P. leei, P. lifesoni, and P. pearti. Pseudotrichonympha leei and P. lifesoni, both from Coptotermes, are closely related to other Coptotermes symbionts including the type species, P. hertwigi. Pseudotrichonympha pearti is the outlier of the trio, more distantly related to P. leei and P. lifesoni than they are to one another, and contains unique features, including an unusual rotating intracellular structure of unknown function.
Introduction
Pseudotrichonympha is a genus of parabasalian protists found exclusively in the hindguts of lower termites, in particular rhinotermitids, where they play a key role in a well-studied symbiotic system in which the microbial community degrades the lignocellulose that makes up most of the animal’s diet1. The ancestral parabasalian body plan was probably a relatively simple cell, characterized by the presence of a hydrogenosome, an anaerobic metabolic organelle derived from the mitochondrion, and akaryomastigont system comprising a nucleus, four flagella, and other conserved cytoskeletal elements2. But the morphology of parabasalians diversified greatly within the context of symbiosis with insects: cells expanded in size, and in some lineages cytoskeletal elements were replicated to form patterns both so grand and complex that they were considered to form a lineage, the so-called hypermastigotes3. Molecular phylogeny and morphology have now shown this elaboration of form actually happened independently in several parabasalian subgroups4. Pseudotrichonympha is a member of one such group, the Trichonymphida, which are characterized by very large cells with a single nucleus that are covered by thousands or tens of thousands of flagella, typically organized in longitudinal rows that extend over most of the cell body and emerge from an anterior organizing centre beneath an apical cap. Other hypermastigotes can be found in the class Cristamonadea, which typically have multiple karyomastigonts rather than a single nucleus (with exceptions such as Deltotrichonympha and Kofoidia), and in the class Spirotrichonymphea, where flagellar bands are organized in a spiral array consisting of multiple right-handed helices.
The overall body plan of Pseudotrichonympha matches the main characteristics of Trichonymphida as a whole, and is specifically distinguished by a robust apical cap, a sub-apical rostrum with an anterior band of short flagella and a posterior band of distinctively long flagella, and a main body that is typically elongated and covered with shorter flagella organized in longitudinal rows (which may have a slight spiral on close inspection). Pseudotrichonympha occurs in many different termite host species, but its diversity and taxonomy are both understudied, and a long history of mistaken identities has led to too much contradictions and confusion in its classification. The genus was first described over a century ago, but was initially and erroneously interpreted as one of the ‘sexes’ of Trichonympha 5. This was corrected by the creation of the new genus, with the type species Pseudotrichonympha hertwigi 6. The type species has since been re-examined by light and electron microscopy and its small subunit rRNA gene sequences (SSU rRNA) characterized for phylogenetic analysis7. But most of the diversity of Pseudotrichonympha remains unexamined. They are known to be common, perhaps ubiquitous8, in the rhinotermitids, a widely-distributed and speciose family of termites, but only about a dozen species have been formally described7. Their diversity is also evident from molecular surveys of termite hindgut communities, which have yielded a diverse clade of environmental sequences that are inferred to be from Pseudotrichonympha, but currently sequences from organisms lacking formal taxonomic description outnumber sequences from described species by a ratio of five to one7. Here, we describe three new species of Pseudotrichonympha from barcoded Coptotermes and Prorhinotermes hosts from North America and Australia, using single cell isolation to provide molecular data from the SSU rRNA for phylogenetic analysis so as to further fill the spaces between identified species and environmental sequences in the Pseudotrichonympha tree.
Results and Discussion
Host collection and identification
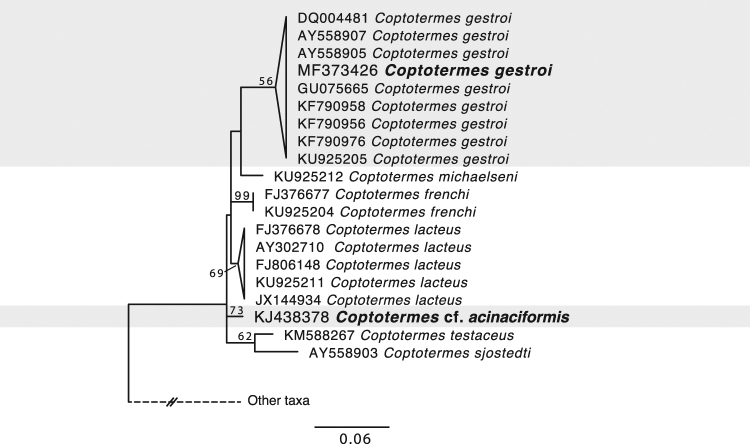
Prorhinotermes simplex and Coptotermes gestroi were collected in Fort Lauderdale, Florida, USA, and their morphological identification was confirmed by DNA barcoding (accessions JX975355 and MF373426, respectively). A second species of Coptotermes was collected in Mount Glorious, Queensland, Australia. It was identified to the genus level by morphology in the field. Ethanol-preserved samples were returned to the lab for DNA barcoding using the mitochondrial large subunit rRNA gene (accession KJ438378), which allowed for comparisons with two of the three species of Coptotermes known from the area. Coptotermes frenchi barcodes shared on average 95.5% identity with the Australian barcode and showed no close affinity in the phylogenetic tree (Fig. 1, Suppl. Figure 1). The identity shared with Coptotermes lacteus barcodes was on average 96.7%, and in phylogenetic trees the Australian barcode branched closer to C. lacteus, but did not branch within the clade of C. lacteus barcode isolates (which share 99–100% identity). Based on this, we conclude that the collection corresponds to neither C. lacteus nor C. frenchi. The only other Coptotermes known in this region is Coptotermes acinaciformis, but unfortunately there is no barcoded vouchered specimen of this species (Coptotermes are notorious difficult to identify). Therefore, we conclude that the probable identity of our isolate is C. acinaciformis, but it could also be a presently unknown species. We will henceforth refer to this specimen as Coptotermes cf. acinaciformis.
Figure 1.
Maximum likelihood phylogeny of host barcode sequences (mitochondrial LSU rRNA) from Coptotermes. All available Coptotermes barcodes are included. The new specimen from Florida is confirmed to correspond to C. gestroi, while the new specimen from Australia is a species for which no barcode is currently available. Based on the available barcodes to which it does not correspond and the diversity of Coptotermes from this region of Queensland, we infer this is most likely a specimen of C. acinaciformis, and therefore refer to this as Coptotermes cf. acinaciformis. Conspecific clades are collapsed and represented by triangles. Numbers at nodes correspond to ML bootstrap support over 50% (values for nodes with lower support are not shown for clarity), and the scale bar represents a distance of 0.06 substitutions per site. The complete tree with outgroups can be seen in Suppl. Figure 1.
Morphology of new Pseudotrichonympha species
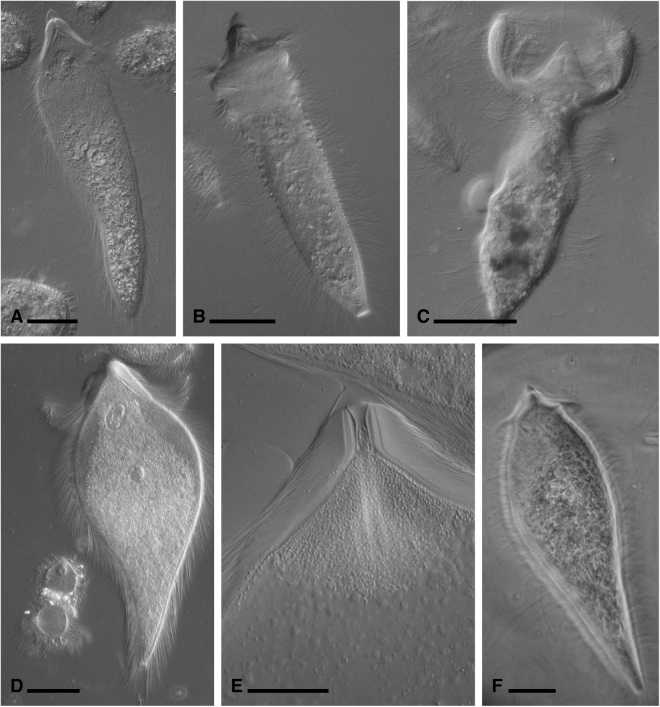
In all three termite species we observed and photographed cells matching the description of Pseudotrichonympha (Fig. 2). Specifically, all three symbionts were characterized by an apical cap with an adjacent apical band of short rostral flagella, a distinctive posterior band of very long rostral flagella, and intermediate-length flagella covering the remainder of the post-rostral body. Cells contained a single nucleus central in the sub-rostral area (e.g. see Fig. 2E). The Pseudotrichonympha from Coptotermes cf. acinaciformis (Fig. 2F) was very large (length average: 392 µm, length range: 332–448 µm, width average: 100 µm,width range: 94–108, n = 5) and had a wide ovoid shape and a sharply tapering posterior tip. The Pseudotrichonympha from C. gestroi (Fig. 2A–C) had a more slender body (length average: 303 µm, length range: 280–330 µm, width average: 62 µm,width range: 56–71 µm, n = 5), and was distinguished by the tendency to possess a robust sub-apical shoulder region that frequently had the greatest diameter of the body (Fig. 2B). In stressed cells (e.g. during isolation), this region expanded greatly and produced a distinctive cup-shaped cell with the apex at the centre of the cup (Fig. 2C). The Pseudotrichonympha from P. simplex (Fig. 2D,E) tended to have a wide body around the midline (length average: 262 µm, length range: 200–316 µm, width average: 122 µm, width range: 89–180 µm, n = 12), and was most obviously defined by the presence of a strange intracellular body we dubbed the “rotatosome”.
Figure 2.
Light micrographs of new Pseudotrichonympha specimens. (A–C) Differential interference contrast images of Pseudotrichonympha leei from Coptotermes gestroi showing the overall body shape, with the long posterior rostral flagella emerging below the pointed apical cap, and shorter post-rostral flagella (A). (B,C) Show the commonly observed robust collar (B) and cup-shaped anterior of stressed cells (C). (D,E) Differential interference contrast images of Pseudotrichonympha pearti from Prorhinotermes simplex showing the overall body shape, single nucleus, and detail of the rostral region, including the apical cap, rostral tube, and long posterior rostral flagella. (F) Phase contrast images (taken in the field) of Pseudotrichonympha lifesoni from Coptotermes cf. acinaciformis showing the basic body shape and size and the single central nucleus. Bars represent 50 µm.
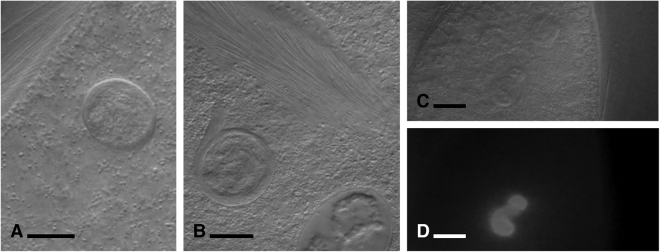
The rotatosome (see Fig. 3, and Supplemental video files) is a spherical body on average 13 µm in diameter (n = 3) that was only ever observed in a single copy per cell. It could only be observed in some focal planes, so whether it is in all cells or not is not known. It continuously rotated, causing visible turbulence in the cytoplasm around it. The lumen of the rotatosome was not uniform. At certain observational planes the rotatosome could be seen to include a rod 6-7 µm in length that emerged from one side of the circle (which side depended on the angle from which it was viewed), was stationary with respect to the cytoplasm, and in which the material appears to stream towards the outermost layer of the sphere, where it can then be seen to turn and turn again in the direction of the rotation. We examined the possibility that the rotatosome is an endosymbiont living within Pseudotrichonympha by Hoechst staining. In fixed and stained cells, the nucleus consistently showed a strong signal for the presence of DNA (in the pattern expected for a parabasalian, which have condensed interphase chromosomes), but the rotatosome retained no detectible stain (Fig. 3C,D). Similarly, we manually broke three Pseudotrichonympha cells open under the microscope to see if a rotatosome could be observed swimming freely, but the body consistently disappeared when cells were disrupted (not shown). A spherical structure was previously observed, in 1923, in the cytoplasm of a species identified as P. sphaerophora from a termite identified as Rhinotermes nasutus in British Guiana9. In this case, the sphere was reported to be larger (25 µm), consisting of a non-staining centre covered by a lighter layer in a concentric organization. The structured was reported from a fixed and stained sample, so no rotation was observed, and it was hypothesized at the time to function as a stercoma for storing excretory material. Based on the difference in size, lack of detail in the 1923 description, and the fact this is apparently not the same species of host or symbiont, or the same collection location, it would be premature to conclude these are the same structure. Overall, in either case, many questions remain about the function and the form of the rotatosome that will hopefully be resolved in the fullness of time, but it appears to be a sub-cellular structure rather than an endosymbiont.
Figure 3.
Details of the “rotatosome” from Pseudotrichonympha pearti. Differential interference contrast details of the rotating structure in the P. pearti cytoplasm (A,B), with (B) showing the projection into the cytoplasm (this can also be seen in the Supplemental video, which also shows how the structure rotates). (C and D) show a rotatosome next to the P. pearti nucleus in fixed and Hoechst stained cells. In C (the DIC image) both structures are visible, but in D (Hoechst stain fluorescence) only the nucleus can be seen to fluoresce, showing the rotatosome does not contain DNA. Bars represent 10 µm.
Molecular phylogeny
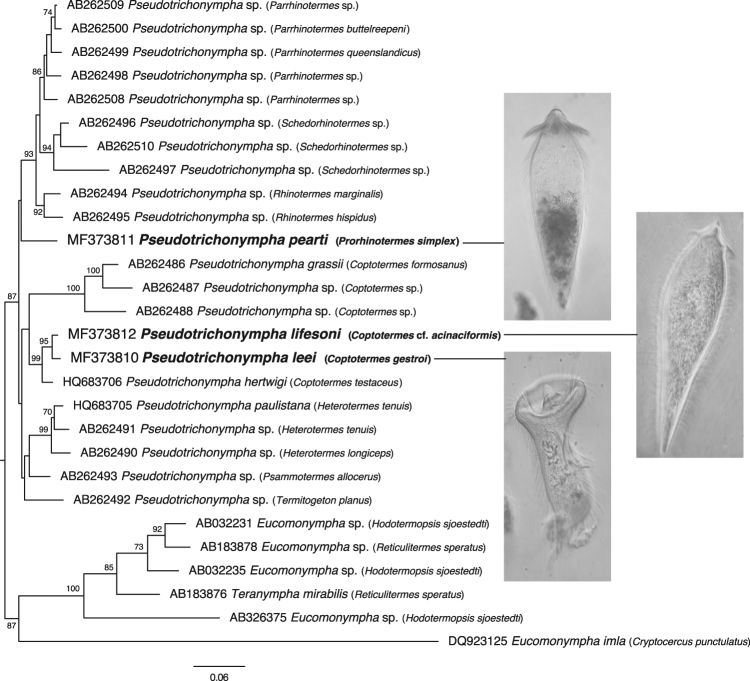
To determine how these Pseudotrichonympha species relate to other members of the genus, we sequenced the SSU rRNA gene from single isolated cells and small pools of cells. In all cases, sequences from the symbionts of a given host were 98.5–99% identical and one representative sequence per taxon was used for phylogenetic analyses and submitted to GenBank under accessions MF373810 –MF373812. In phylogenetic trees, the Pseudotrichonympha symbionts from Coptotermes hosts branch within a single clade, albeit with no support (Fig. 4). However, within this clade are two well-supported subgroups, one of these consists of three sequences from Southeast Asian specimens of Coptotermes (and includes one known species, P. grassii from C. formosanus), and the other consists of P. hertwigi from its type host, C. testaceus, and the two Coptotermes symbionts described here. The P. simplex symbiont is excluded from this clade, but its position is otherwise unresolved and the real relation to the underlying tree of currently sampled Pseudotrichonympha sequences unclear (Fig. 4).
Figure 4.
Maximum likelihood (ML) phylogeny of SSU rRNA genes from Pseudotrichonympha, showing the relationships between Pseudotrichonympha leei, Pseudotrichonympha lifesoni, and Pseudotrichonympha pearti, to other members of the genus. The tree is rooted with the closest known relatives to Pseudotrichonympha, Teranympha and Eucomonypha. Numbers at nodes correspond to ML bootstrap support over 70% (values for nodes with lower support are not shown for clarity), and the scale bar represents a distance of 0.06 substitutions per site. Taxon names include GenBank accession numbers and the name of the termite host, where known. Most of the diversity on the phylogeny is represented by sequences from undescribed species, many from unidentified hosts, emphasizing our dearth of knowledge about the diversity of this lineage.
Taxonomic considerations
All three specimens of Pseudotrichonympha characterized here represent new species. In the case of C. gestroi, flagellates have been reported10 but none, including members of the genus Pseudotrichonympha, have been formally described. In the case of P. simplex, several spirotrichonymphids and the smaller flagellate, Cthulhu, have been described11, but although Pseudotrichonympha has been reported to be present, it was never formally described10–12.
In the case of the Australian Coptotermes cf. acinaciformis, we can conclude the symbiont is undescribed without knowing the host species with absolute certainty. Of the three Coptotermes known to exist in the region, one (C. frenchi) has no described Pseudotrichonympha, and the other two (C. lacteus and C. acinaciformis) are both documented as containing “Pseudotrichonympha hertwigi”10,13. However, this host barcode excludes both C. frenchi and C. lacteus, and moreover the confusing history of P. hertwigi is inconsistent with the presence of this species in any of these hosts (reviewed in detail in ref.7). Since its original description and subsequent re-classificiation to a new genus5,6,14, several authors have assumed that Pseudotrichonympha observed in other Coptotermes hosts correspond to the same species, even in geographically distant Australian and Asian termites10,12,13. But this was invariably based only on light microscopy and limited numbers of distinguishable characters. The identity of the host species is also convoluted. The Coptotermes host of P. hertwigi was named Coptotermes hartmanni 15, but it lacked formal description and is now invalid16. Subsequent surveys of neotropical Coptotermes have shown all Brazilian specimens correspond to Coptotermes testaceus: this host has now been barcoded and the P. hertwigi SSU rRNA from this type host has been characterized7. This species does not exist in Australia, and neither the Pseudotrichonympha from the Australian Coptotermes nor the host barcodes match their Brazilian counterparts, leading to the conclusion that neither Australian C. lacteus nor C. acinaciformis (nor any other Australian termite) contain P. hertwigi, but rather contain undescribed species that have been misidentified. The same is also likely true for Asian termites concluded by light microscopy alone to harbour P. hertwigi. Based on this and the phylogenetic and morphological data, we propose three new species of Pseudotrichonympha as detailed below.
Taxonomic Summary
Pseudotrichonympha leei n. sp. del Campo and Keeling, 2017
urn:lsid:zoobank.org:act:33F52F74-954F-4F0F-9726-112C7D761266
Type host: Coptotermes gestroi (Isoptera, Rhinotermitidae: barcode MF373426).
Type locality: Ft. Lauderdale, Secrete Woods County Park, Florida, USA: lat. 26.08567, long. -80.18017.
Host collection: University of Florida termite collection, accession number FL3578. Collector R. H. Scheffrahn. Collected April 8, 2011.
Description: Parabasalian flagellate with morphological characteristics of the genus Pseudotrichonympha. Cells are on average 303 µm in length (range: 280–330 µm, n = 5) and 62 µm in width (range: 56–71 µm, n = 5). Cells are typically narrow and elongated, and can form a pronounced subapical collar, just posterior to the rostral flagella, which can, under stress, extend to give the cell a distinctive cup-shaped apex. Found in the hindgut of Coptotermes gestroi. Distinct SSU rRNA sequence (GenBank accession number MF373810).
Holotype: Specimen in Fig. 1A of the present publication.
Gene sequence: SSU rRNA accession number MF373810.
Etymology: Species name refers to the Geddy Lee, a musician who, with other members of Rush, have inspired an interest in natural history and science through art.
Pseudotrichonympha lifesoni n. sp. del Campo and Keeling, 2017
urn:lsid:zoobank.org:act:C214D2FD-196A-4842-BCFF-9DA60F48B862
Type host: Coptotermes cf. acinaciformis (Isoptera, Rhinotermitidae: barcode KJ438378).
Type locality: Mount Glorious, 1780 Mt. Glorious Road, Queensland, Australia: lat. -27.33773, long. 152.77028.
Host collection: University of Florida termite collection, accession number AUS115. Collector P. J. Keeling. Collected Nov. 17, 2011.
Description: Parabasalian flagellate with morphological characteristics of the genus Pseudotrichonympha. Cells are on average 392 µm in length (range: 332–448 µm, n = 5) and 100 µm in width (range: 94–108 µm, n = 5). Cells are ovoid with a strongly elongated posterior tip and a clearly demarcated rostral flagellar zone. Found in the hindgut of Coptotermes cf. acinaciformis with a distinct barcode identification (KJ438378). Distinct SSU rRNA sequence (GenBank accession number MF373812).
Holotype: Specimen in Fig. 1F of the present publication.
Gene sequence: SSU rRNA accession number MF373812.
Etymology: Species name refers to the Alex Lifeson, a musician who, with other members of Rush, have inspired an interest in natural history and science through art.
Pseudotrichonympha pearti n. sp. del Campo and Keeling, 2017
urn:lsid:zoobank.org:act:907E8D63-BA08-4B37-A36A-EDD7572D9A5E
Type host: Prorhinotermes simplex (Isoptera, Rhinotermitidae: barcode JX975355).
Type locality: Ft. Lauderdale, Secrete Woods County Park, Florida, USA: lat. 26.08567, long. -80.18017.
Host collection: University of Florida termite collection, accession number FL1563. Collector R. B. Maharajh. Collected Sept. 15, 2002.
Description: Parabasalian flagellate with morphological characteristics of the genus Pseudotrichonympha. Cells are on average 262 µm in length (range: 200–316 µm, n = 12) and 122 µm in width (range: 89–180 µm, n = 12). Cells contain a distinctive single, rotating spherical body, the rotatosome, with a diameter of approximately 13 µm. Found in the hindgut of Prorhinotermes simplex. Distinct SSU rRNA sequence (GenBank accession number MF373811)
Holotype: Specimen in Fig. 1D of the present publication.
Gene sequence: SSU rRNA accession number MF373811.
Etymology: Species name refers to the Neil Peart, a musician who, with other members of Rush, have inspired an interest in natural history and science through art.
Methods
Host termite collection and barcoding
Prorhinotermes simplex and Coptotermes gestroi were collected in Secret Woods County Park, Fort Lauderdale, Florida, USA, on September 15, 2002 and April 8, 2011, respectively. A Coptotermes that was identified morphologically to the genus level (probably C. acinaciformis) was collected at the Turkey Nest, Mount Glorious, Queensland, Australia, Nov. 17, 2011. All specimens were deposited in the University of Florida termite collection under accessions FL1563, FL3578, and AUS115, respectively. Prorhinotermes simplex and Coptotermes gestroi were maintained in conical tubes with wood from their habitats at room temperature in the laboratory. Coptotermes cf. acinaciformis was processed directly in the field. Termite identities were determined morphologically and by barcoding using the mitochondrial 16 S (LSU) ribosomal RNA gene amplified and sequenced using the primers LR-N-13398 5′-CGC CTG TTT ATC AAA AAC AT-3′17 and LR-J-13017 5′-TTA CGC TGT TAT CCC TAA-3′18. These were aligned with Coptotermes LSU termite barcodes from Genbank using using MAFFT v7.31019 (setting: –auto). Poorly aligned regions were automatically removed with trimAl v1.4 using a gap threshold of 0.320 (settings: –gt 0.3 –st 0.001); columns with missing data at both ends were removed. For the termite phylogeny, maximum likelihood (ML) inference was carried out using RAxML v8.2.9 assuming the GTR + Γ substitution model with 1,000 random starting trees and statistical support obtained from 1,000 bootstrap replicates21. Barcodes were submitted to GenBank under accessions JX97535511, KJ43837822 and MF373426 (this study).
Microscopy
Termites were dissected and hindgut contents were suspended in Trager’s medium U23. Symbionts from C. gestroi and P. simplex were observed on an Axioplan 2 compound microscope (Zeiss, Oberkochen, Germany) using differential interference contrast and documented with a 3CCD HD video camera XL H1S (Canon, Tokyo, Japan). Multiple cells of each new species were observed, filmed, and measured to estimate ranges of cell size and shape, but in all cases the size range was variable and many cells were observed to deviate from the norm, as these species are known to display significant plasticity. For this reason, we also carried out molecular characterization using single cell isolations of these symbionts, using an Axiovert 200 (Zeiss, Oberkochen, Germany) inverted microscope. Isolated cells were photographed with a MicroImager II (QImaging, Surrey, BC, Canada). For Coptotermes cf. acinaciformis, microscopy was carried out at the sampling location using a Swift Field Master phase contrast field microscope (Swift Optical Instruments, Schertz, TX) and imaging using a Canon G9 camera (Canon, Tokyo, Japan). Whether the “rotatosome” structure was an endosymbiotic cell was tested by puncturing individual cells with a small-diameter micro-pipette. We observed that with a certain amount of force the cell membrane could be disrupted without damaging comparably sized organelles, such as the nucleus.
For fluorescence microscopy, cells from P. simplex were fixed and stained in 2% paraformaldehyde in PBS buffer (pH 7.4) with 10 μg/mL Bisbenzimide H 33258 (Hoescht stain: Sigma, St Louis, MO) for 10 min. The cells were observed under an Axioplan2 fluorescent microscope and filmed with the Canon XL H1S. Bisbenzimide fluorescence were detected with the UV filter (excitation BP 365/12 nm, emission LP 397 nm).
Single cell isolation, sequencing, and phylogenetic analysis
Individual Pseudotrichonympha cells were isolated by glass micropipette. Symbionts from P. simplex and C. gestroi were isolated in the lab using a Zeiss Axiovert, while individual Pseudotrichonympha cells from Coptotermes cf. acinaciformis were isolated directly in the field using the Swift Field Master. Representatives of this species were fixed in 95% ethanol with a view towards its preservation and shipped to the University of British Columbia for all subsequent molecular analyses. For all three species, single cells or small pools of cells were manually picked and rinsed three times under constant observation. In cases where the cell was so close to the edge of the cavity slide as to obscure the view, they were washed an additional time and inspected to confirm its state of integrity was not compromised. DNA was extracted from isolated cells using the Masterpure Complete DNA and RNA Purification Kit (Epicentre, Madison, WI, USA). SSU rRNA genes were amplified from purified DNA using the eukaryote specific primers PFI 5′-TGC GCT ACC TGG TTG ATC CTG CC-3′ and FAD4 5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′. PCR conditions included a 3-minute denaturation at 95 °C followed by 30 cycles of 95 °C for 30 seconds, 55 °C for 30 seconds, and 72 °C for 1 minute 30 seconds, then an additional 7 minutes at 72 °C. Products were purified, cloned into the pCR2.1 vector using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA, USA), and sequenced on both strands with BigDye Terminator v 3.1 (Applied Biosystems, Carlsbad, CA, USA). Multiple clones were sequenced from each isolation. Sequences were submitted to GenBank under accessions MF373810 - MF373812.
New Pseudotrichonympha sequences were aligned with previously published sequences spanning the phylogenetic diversity of parabasalians using MAFFT with default settings19. Highly variable regions were removed using trimAl20 (settings: –gt 0.3 –st 0.001). Preliminary analyses confirmed with complete support that all new sequences corresponded to Pseudotrichonympha, and a more detailed analysis focused exclusively on sequences from this genus and outgroups, resulting in a final alignment of 28 taxa and 1,388 positions. ML analyses were performed with RAxML v8.2.921, using the GTR + Γmodel and 1,000 random starting trees. For the ML analysis, support was assessed from 1,000 bootstrap replicates.
Electronic supplementary material
Acknowledgements
We thank Phil Hugenholz for assistance with termite collections in Australia, the owners of the Turkey Nest Cabin for their hospitality and permission to explore their land for all the busy little creatures we sought, and the members of Rush for forty years. This work was supported by a grant (RGPIN-2014-03994) from the Natural Sciences and Engineering Research Council of Canada to PJK. JdC, YH, VB, AK, EH and MK were supported by grants to the Centre for Microbial Diversity and Evolution from the Tula Foundation, and NATI was supported by a fellowship from NSERC. JdC was supported by a Marie Curie International Outgoing Fellowship grant (FP7-PEOPLE-2012-IOF - 331450 CAARL).
Author Contributions
P.J.K. and J.d.C. wrote the manuscript. R.H.S. and P.J.K. retrieved the termite specimens. E.R.J. and P.J.K. isolated the parabasalian cells. J.d.C., E.R.J., Y.H., R.F., M.K., N.A.T.I., V.M., V.B., E.H. and A.K. retrieved and analyzed the molecular and imaging data. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16259-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 2014;12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 2.Brugerolle, G. & Lee, J. J. Parabasalia in The IllustratedGuideto the Protozoa (eds. Lee, J. J., Leedale, G. F. & Bradbury, P. C.) 1196–1250 (Society of Protozoology, 2000).
- 3.Hollande A, Carruette-Valentin J. Les atractophores, l’induction du fuseau et la division cellulaire chez les Hypermastigines. Étudeinfrastructurale et révisionsystématique des Trichonymphines et des Spirotrichonymphines. Protistologica. 1971;7:5–100. [Google Scholar]
- 4.Čepička I, Hampl V, Kulda J. Critical taxonomic revision of parabasalids with description of one new genus and three new species. Protist. 2010;161:400–433. doi: 10.1016/j.protis.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Hartmann M. UntersuchungenüberBau und Entwicklung der Trichonymphiden (Trichonymphahertwigi n. sp.) Festschrift SechzigstenGeburtstag Richard Hertwigs. 1910;1:351–396. [Google Scholar]
- 6.Grassi B, Foà A. Intornoaiprotozoideitermitidi. Rend. R. Accad. Lincei, ser. 5, sem. 1911;112:725–741. [Google Scholar]
- 7.Saldarriaga JF, et al. Morphology and molecular phylogeny of Pseudotrichonymphahertwigi and Pseudotrichonymphapaulistana (Trichonymphea, Parabasalia) from Neotropical Rhinotermitids. J. Eukaryot. Microbiol. 2011;58:487–496. doi: 10.1111/j.1550-7408.2011.00575.x. [DOI] [PubMed] [Google Scholar]
- 8.Noda S, et al. Cospeciation in the triplex symbiosis of termite gut protists (Pseudotrichonympha spp.), their hosts, and their bacterial endosymbiosis. Mol. Ecol. 2007;16:1257–1266. doi: 10.1111/j.1365-294X.2006.03219.x. [DOI] [PubMed] [Google Scholar]
- 9.Dunkerley JS. A new structure in the flagellate Pseudotrichonymphasphaerophora sp. n. Parasitology1. 1923;5:211–212. doi: 10.1017/S0031182000014670. [DOI] [Google Scholar]
- 10.Yamin MA. Flagellates of the orders Trichomonadida Kirby, OxymonadidaGrassé, and HypermastigidaGrassi&Foàreported from lower termites (Isoptera families Mastotermitidae, Kalotermitidae, Hodotermitidae, Termopsidae, Rhinotermitidae, and Serritermitidae) and from the wood-feeding roach Cryptocercus (Dictyoptera: Cryptocercidae) Sociobiology. 1979;4:5–119. [Google Scholar]
- 11.James ER, Okamoto N, Burki F, Scheffrahn RH, Keeling PJ. Cthulhu macrofasciculumque n. g., n. sp. and Cthyllamicrofasciculumque n. g., n. sp., a newly identified lineage of parabasalian termite symbionts. PLoS ONE. 2013;8:e58509. doi: 10.1371/journal.pone.0058509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleveland LR. Symbiosis between termites and their intestinal protozoa. Proc. Natl. Acad. Sci. USA. 1923;9:424–428. doi: 10.1073/pnas.9.12.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannesmann RVU. den Einflußder Temperatur auf die Darmsymbionten von Termiten und über die regulatorischenMechanismen der SymbioseTeil 1. Zeitschrift fur angewandteZoologie. 1969;56:385–440. [Google Scholar]
- 14.Grassi B. FlagellativiventineiTermiti. Rend. R. Accad. Lincei, ser. 1917;512:331–394. [Google Scholar]
- 15.Holmgren N. Termitenstudien. 2. Systematik der Termiten: die FamilienMastotermitidae, Protermitidae und Mesotermitidae. K. SvenskaVetensk. Akad. Handl. 1911;46:1–88. [Google Scholar]
- 16.Snyder TE. Catalog of the termites (Isoptera) of the world. Smithson. Misc. Collect. 1949;112:1–490. [Google Scholar]
- 17.Simon C, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- 18.Kambhampati S, Smith PT. PCR primers for the amplification of four insect mitochondrial gene fragments. Insect Mol. Biol. 1995;4:233–236. doi: 10.1111/j.1365-2583.1995.tb00028.x. [DOI] [PubMed] [Google Scholar]
- 19.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tai V, et al. The role of host phylogeny varies in shaping microbial diversity in the hindguts of lower termites. Appl. Env. Microbiol. 2015;81:1059–1070. doi: 10.1128/AEM.02945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trager W. The cultivation of a cellulose-digesting flagellate, Trichomonas termopsidis, and of certain other termite protozoa. Biol. Bull. 1934;66:182–190. doi: 10.2307/1537331. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.