Abstract
Genome-wide association studies (GWASs) have been performed extensively in diverse populations to identify single nucleotide polymorphisms (SNPs) associated with complex diseases or traits. However, to date, the SNPs identified fail to explain a large proportion of the variance of the traits/diseases. GWASs on type 2 diabetes (T2D) and obesity are generally focused on individual traits independently, and genetic intercommunity (common genetic contributions or the product of over correlated phenotypic world) between them are largely unknown, despite extensive data showing that these two phenotypes share both genetic and environmental risk factors. Here, we applied a recently developed genetic pleiotropic conditional false discovery rate (cFDR) approach to discover novel loci associated with BMI and T2D by incorporating the summary statistics from existing GWASs of these two traits. Conditional Q-Q and fold enrichment plots were used to visually demonstrate the strength of pleiotropic enrichment. Adopting a cFDR nominal significance level of 0.05, 287 loci were identified for BMI and 75 loci for T2D, 23 of which for both traits. By incorporating related traits into a conditional analysis framework, we observed significant pleiotropic enrichment between obesity and T2D. These findings may provide novel insights into the etiology of obesity and T2D, individually and jointly.
Introduction
Genome-wide association studies (GWASs) have successfully identified hundreds of SNPs associated with complex diseases or traits. However, to date, the SNPs identified fail to explain a large proportion of the variance of the traits/diseases under study. Previous studies have suggested that GWASs have the potential to explain a larger proportion of “missing heritability”1,2 mainly by using larger sample sizes3. However, although acquiring larger sample sizes may increase statistical power, it is often not feasible since the recruiting and genotyping of additional participants is too costly. Therefore, there is a need for analytical methods that can better and more efficiently utilize the information contained in the existing pool of available data for the identification of trait-associated loci. Several of these types of methods have recently been developed4–6 and successfully applied7,8 to identify novel loci for various complex traits.
Pleiotropy is the phenomenon of a single gene or locus affecting two or more phenotypes9. There is ample evidence to suggest that genetic pleiotropy exists in many correlated diseases and traits, such as bipolar disorder and schizophrenia10, indicating that related traits may share overlapping genetic mechanisms. Through the incorporation of information regarding genetic pleiotropy, we can improve the detection power of common variants associated with complex diseases or traits by effectively increasing the sample sizes without the need to recruit more individuals. The joint analysis of related phenotypes may reveal novel insights into the common biological mechanisms and overlapping pathophysiological relationships between complex traits.
Andreassen et al.4 developed a genetic-pleiotropy-informed conditional false discovery rate (cFDR) method by leveraging two GWASs from associated traits in a conditional analysis. The method has been successfully applied to genetically associated diseases and phenotypes including schizophrenia and bipolar disorder7, as well as blood pressure and other phenotypes8. Our group has recently successfully applied the cFDR method to the joint analyses of bone mineral density (BMD) and breast cancer11, BMD and coronary artery disease12, femoral neck (FNK) BMD and height13, and T2D and birth weight14. All of these studies improved statistical power through the joint analysis of related traits, and unambiguously demonstrated the utility of the method for improving the identification of potentially novel trait-associated variants.
Obesity is a chronic metabolic disorder mainly characterized by excessive body fat. Body Mass Index (BMI) is widely used in obesity research and clinical diagnosis to quantify an individual’s tissue mass. Identification of the genetic determinants for BMI, a non-invasive measure of obesity that predicts the risk of related complications15, could lead to a better understanding of the biological basis of obesity. Epidemiological studies estimate that the prevalence of overweight/obese individuals increased by >40% between 1980 and 201316, and that these elevated obesity levels are a driving force for the similarly rapid increase of Type II Diabetes (T2D)17. T2D is a chronic metabolic disorder characterized by high blood sugar, insulin resistance, and relative lack of insulin, all of which share some genetic susceptibility and functional mechanisms with obesity18. Heritability studies have demonstrated a substantial genetic contribution to both obesity risk (h2~40–70%)19 and T2D (h2~26–69%)20. In 2014, an estimated 387 million people were living with diabetes, corresponding to a worldwide prevalence of 8.3%, and 90% of these individuals had T2D21.
There has been substantial evidence to indicate an important relationship between obesity and T2D, along with strong support22,23 to suggest that obesity and T2D share some common genetic risk factors. The accumulation of body fat may be associated with several conditions related to T2D including insulin resistance, hyperinsulinemia, the reduced utilization of glucose in muscles and other tissues, and impaired glucose tolerance24. Additionally, Corbin et al.25 used Mendelian Randomization (MR) Egger analysis to explore the complex relationship between these traits and demonstrated a true causal effect of BMI on T2D, as well as potential pleiotropy between the two phenotypes. Although dozens of genetic loci associated with BMI or T2D have been detected by GWASs26,27, these loci can explain at best 10% of the genetic variance for either obesity28 or T2D29. Considering the high degree of heritability and potential pleiotropy between these phenotypes, the two traits are ideal for the further analyses using the cFDR approach to improve the detection of loci associated with obesity and/or T2D.
In this study, we applied the genetic-pleiotropy-informed cFDR method4 on two large datasets of GWAS summary statistics for BMI and T2D30,31 to identify novel loci and pleiotropic relationships between these traits. These two GWASs have identified 97 and 62 loci associated with BMI and T2D respectively, but they only explain 2.7% and 5.7% of the total heritability for these traits30,31. The purpose of our study is to improve SNP detection for obesity and T2D using these two existing GWASs, and gain some novel insights into the shared biological mechanisms and overlapping genetic heritability between them. The clarification of potentially shared genetic determinants may have significant implications for the identification of important biomarkers and development of novel therapeutic approaches for joint prediction, prevention, and intervention of the two related diseases/phenotypes.
Results
Assessment of pleiotropic enrichment
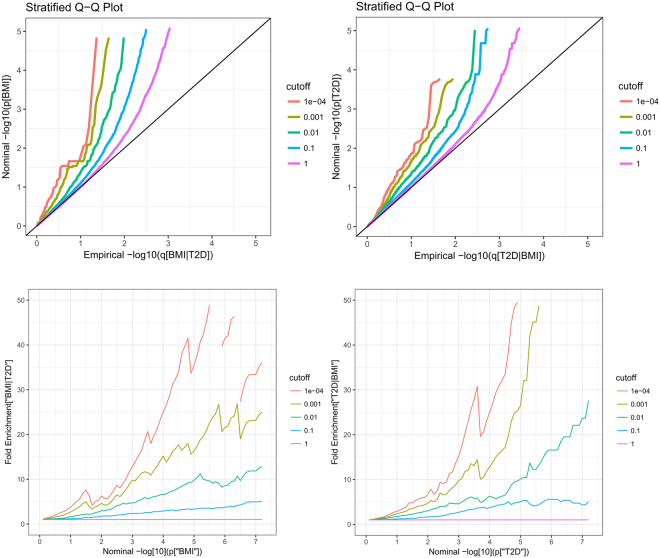
The conditional Q-Q plot for BMI conditional on T2D (Upper Panel (left) in Fig. 1) showed some enrichment across varying significance thresholds for T2D. The presence of leftward shift when restricting the analysis to include the SNPs that have more significant associations with BMI indicates an increase in the number of true associations for a given T2D p-value. Similar enrichment is observed for T2D given BMI (Upper Panel (right) in Fig. 1), as there appears to be a similar departure pattern between the different curves. These leftward deflections from the null line indicate a greater proportion of true associations for any given BMI nominal p-value.
Figure 1.
Stratified QQ (upper panel) and Enrichment (lower panel) plots. Upper Panel: Stratified QQ plots of nominal versus empirical –log10 p-values in (left) BMI as a function of significance of the association with T2D, and in (right) T2D as a function of significance of the association with BMI. Lower Panel: Fold-enrichment plots of enrichment versus nominal –log10 p-values for (left) BMI below the standard GWAS threshold of p < 5 × 10−8 as a function of significance of the association with T2D, and (right) T2D below the standard GWAS threshold of p < 5 × 10−8 as a function of significance of the association with BMI. The purple line with slope of zero represents all SNPs.
Based on the fold-enrichment plot (Lower Panel of Fig. 1), we observed SNP enrichment for BMI across different levels of significance with T2D and vice versa. For progressively stringent p-value thresholds for BMI SNPs, we observed about a 50-fold increase in the proportion of SNPs reaching the genome wide significance level of -log10 (p) > 7.3 when comparing the subset with the most stringent conditional association to the group with all SNPs. A 50-fold increase was also observed for T2D.
As negative controls, conditional Q-Q plots for BMI given nominal p-values of association with attention-deficit/hyperactivity disorder (ADHD) (Upper Panel (left) in Figure S1) and major depressive disorder (MDD) (Lower Panel (left) in Figure S1), and T2D conditional on ADHD (Upper Panel (left) in Figure S2) and MDD (Lower Panel (left) in Figure S2) all showed no enrichment and vice versa.
BMI loci identified with cFDR
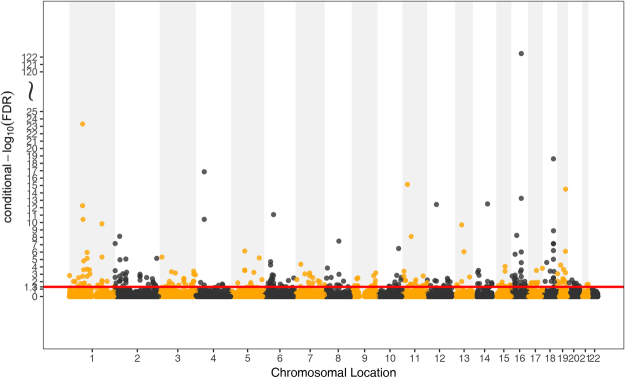
Conditional on their association with T2D, we identified 287 significant SNPs (cFDR < 0.05) for BMI variation (Fig. 2 and Table S2), which were mapped to 21 different chromosomes (1–21) and annotated to 323 genes. In the original meta-analysis for BMI GWAS31, 105 SNPs had p-values smaller than 1 × 10−5 while 36 of them reached the standard genome-wide significance of 5 × 10−8. We confirmed 43 SNPs that were reported in the original BMI GWAS analysis31 and previous BMI related GWASs32. Another 40 SNPs that were reported to be associated with BMI-related traits were also confirmed in our analysis32–34. The rest of the 204 SNPs were not previously reported in the original BMI GWAS31 or any other previous obesity studies. However, 26 of these 204 SNPs are in high linkage disequilibrium (LD) (r2 > 0.6) with other BMI-associated SNPs reported previously (Table S3). Among the 323 genes these 287 SNPs were annotated to, 146 of these genes were newly detected compared to the original BMI GWAS31 and previous obesity-related studies (Table S2). Among all the 287 detected loci for BMI, most of the genes were enriched in BMI-related terms such as “positive regulation of cellular metabolic process”, “positive regulation of metabolic process” and “regulation of protein metabolic process”. GO term enrichment analysis results were detailed in Table 1.
Figure 2.
Conditional Manhattan plot of conditional –log10 FDR values for BMI given T2D (BMI|T2D). The red line marks the conditional –log10 FDR value of 1.3 corresponds to a cFDR < 0.05.
Table 1.
Functional Term Enrichment Analysis.
| Pathway ID | Pathway description | Count in gene set | False discovery rate |
|---|---|---|---|
| BMI GO:0031325 | positive regulation of cellular metabolic process | 58 | 0.00983 |
| GO:0031328 | positive regulation of cellular biosynthetic process | 41 | 0.0119 |
| GO:0007275 | multicellular organismal development | 74 | 0.0172 |
| GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 39 | 0.019 |
| GO:0051173 | positive regulation of nitrogen compound metabolic process | 40 | 0.0197 |
| GO:0010604 | positive regulation of macromolecule metabolic process | 53 | 0.0223 |
| GO:0010628 | positive regulation of gene expression | 38 | 0.0223 |
| GO:0051130 | positive regulation of cellular component organization | 30 | 0.0223 |
| GO:0051254 | positive regulation of RNA metabolic process | 35 | 0.0223 |
| GO:0045859 | regulation of protein kinase activity | 21 | 0.0282 |
| GO:0009893 | positive regulation of metabolic process | 63 | 0.0306 |
| GO:0010557 | positive regulation of macromolecule biosynthetic process | 36 | 0.0316 |
| GO:0051246 | regulation of protein metabolic process | 48 | 0.0343 |
| GO:0051338 | regulation of transferase activity | 24 | 0.0379 |
| GO:0044767 | single-organism developmental process | 77 | 0.0449 |
| GO:0035270 | endocrine system development | 8 | 0.0462 |
| T2D GO:0031016 | pancreas development | 6 | 0.00523 |
| GO:0009749 | response to glucose | 6 | 0.0222 |
| GO:0035270 | endocrine system development | 6 | 0.0222 |
| GO:0061017 | hepatoblast differentiation | 2 | 0.0289 |
| GO:0031018 | endocrine pancreas development | 4 | 0.0393 |
| GO:0000976 | transcription regulatory region sequence-specific DNA binding | 10 | 0.0116 |
| GO:0044212 | transcription regulatory region DNA binding | 11 | 0.0116 |
| GO:0043565 | sequence-specific DNA binding | 12 | 0.0214 |
| GO:0051427 | hormone receptor binding | 5 | 0.0478 |
| BMI and T2D GO:0010883 | regulation of lipid storage | 3 | 0.0231 |
| GO:0070344 | regulation of fat cell proliferation | 2 | 0.007 |
| GO:0007267 | cell-cell signaling | 15 | 8.79e–6 |
| GO:0016055 | Wnt signaling pathway | 10 | 2.38e–6 |
| GO:0198738 | cell-cell signaling by wnt | 10 | 2.38e–6 |
| GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 10 | 3.41e–6 |
| GO:0045444 | fat cell differentiation | 5 | 0.00279 |
| GO:0045598 | regulation of fat cell differentiation | 5 | 0.000541 |
| GO:0015908 | fatty acid transport | 4 | 0.000926 |
| GO:0015909 | long-chain fatty acid transport | 4 | 0.000246 |
| GO:0019395 | fatty acid oxidation | 4 | 0.00268 |
| GO:0030258 | lipid modification | 4 | 0.0485 |
| GO:0034440 | lipid oxidation | 4 | 0.00277 |
| GO:0030308 | negative regulation of cell growth | 7 | 3.11e–5 |
| GO:0045600 | positive regulation of fat cell differentiation | 4 | 0.000457 |
| GO:0010565 | regulation of cellular ketone metabolic process | 4 | 0.0239 |
| GO:0019217 | regulation of fatty acid metabolic process | 4 | 0.00394 |
| GO:0045834 | positive regulation of lipid metabolic process | 4 | 0.0146 |
| GO:0045923 | positive regulation of fatty acid metabolic process | 4 | 0.000242 |
| GO:0046320 | regulation of fatty acid oxidation | 4 | 0.000145 |
| GO:0046321 | positive regulation of fatty acid oxidation | 4 | 2.78e–5 |
| GO:0050872 | white fat cell differentiation | 4 | 4.02e–5 |
| GO:0050873 | brown fat cell differentiation | 4 | 0.000653 |
T2D gene loci identified with cFDR
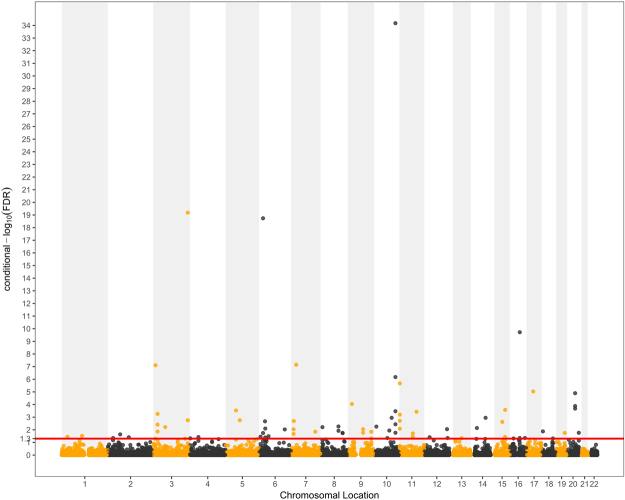
We identified 75 SNPs significantly (cFDR < 0.05) associated with T2D given their association with BMI (Fig. 3 and Table S4), which were located on 20 chromosomes (1–20) and annotated to 89 genes. In the original meta-analysis for T2D GWAS30, 38 SNPs had p-values smaller than 1 × 10−5 while 12 of them reached the standard genome-wide significance of 5 × 10−8. We confirmed 17 SNPs that were reported in the original T2D GWAS analysis30 or previous T2D related GWASs29,35. Another 18 SNPs that were reported to be associated with T2D-related traits were also confirmed in our analysis33,36. The remaining 40 SNPs were not previously reported in the original T2D GWAS30 or any other T2D studies, although nine of these SNPs showed high LD (r2 > 0.6) with the T2D-associated SNPs reported previously (Table S5). For the 89 genes these 75 SNPs were annotated to, 42 of these genes were novel and not identified by the original T2D GWAS30 or previous T2D-related studies (Table S2). Of the detected loci for T2D, some of the genes were enriched in T2D-related terms such as “pancreas development”, “response to glucose” and “endocrine pancreas development”. GO term enrichment analysis were detailed in Table 1.
Figure 3.
Conditional Manhattan plot of conditional –log10 FDR values for T2D given BMI (T2D|BMI). The red line marking the conditional –log10 FDR value of 1.3 corresponds to a cFDR < 0.05.
Pleiotropic gene loci for both BMI and T2D
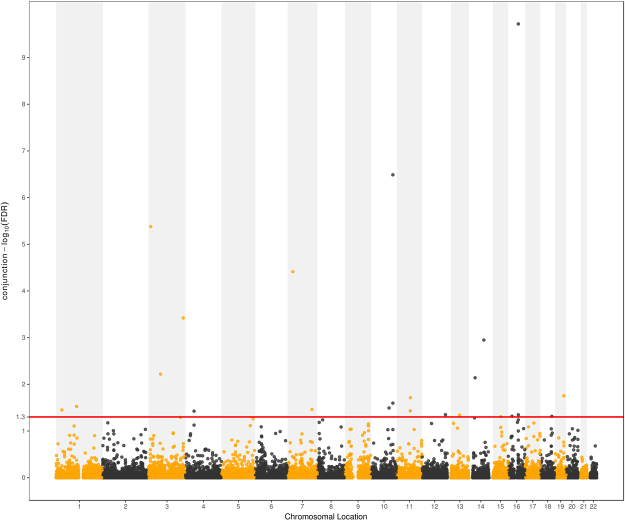
The conjunction FDR analysis detected 23 independent pleiotropic loci that were significantly (conjunction FDR < 0.05) associated with both traits (Fig. 4 and Table 2). Of the 23 identified pleiotropic variants, one SNP rs9930506 (FTO) reached genome-wide significance in the original BMI and T2D GWASs30,31. The SNPs rs7141420 (NRXN3), rs1996023 (GNPDA2 and GABRG1), rs16945088 (FTO), rs9540493 (LOC10272396 and LINC01052) and rs4238585 (GPR139 and GP2) reached genome-wide significance in only the original BMI GWAS31. Six SNPs (rs10787472 (TCF7L2), rs2881654 (PPARG), rs849135 (JAZF1), rs4481184 (IGF2BP2), rs1783598 (FCHSD2) and rs12245680 (TCF7L2)) were reported to be significant for only T2D in the original30 or previous T2D GWAS29. The two SNPs rs6795735 (ADAMTS9-AS2) and rs12454712 (BCL2) were previously reported to be associated with both obesity and T2D29,37,38. The other five SNPs (rs17584208, rs11979110, rs10898868, rs1996023 and rs4474658) were previously reported to be associated with high density lipoprotein (HDL) and proinsulin34,39,40. The final four SNPs were not previously reported in the original BMI and T2D GWASs or GWAS studies for any related traits. For the 30 genes the identified pleiotropic SNPs were annotated to, we found twelve of them (AKAP6, NPAS3, PSRC1, MYBPHL, MIR29A, GABRG1, ZNF664, FAM101A, LOC10272396, LINC01052, GPR139, and PUM1) were not identified by any BMI or T2D related GWASs. For the SNPs that were annotated to these 12 genes, two SNPs were located in the intronic regions of genes ZNF664 and PUM1 respectively, and the other five SNPs were all located in intergenic regions (Table 2). Of the detected 23 pleiotropic loci, most of the genes were enriched in BMI and T2D related terms such as “regulation of lipid storage”, “regulation of fat cell proliferation”, “fat cell differentiation”, and “fatty acid transport”. Detailed information of GO term analysis was given in Table 1.
Figure 4.
Conjunction Manhattan plot of conjunction –log10 FDR values for BMI and T2D. The red line marking the conditional –log10 FDR value of 1.3 corresponds to a conjunction FDR < 0.05. The figure shows the genomic locations of pleiotropic loci and further details are provided in Table 2.
Table 2.
Conjunction FDR: Pleiotropic Loci in BMI and T2D (cFDR < 0.05).
| RSID | ROLE | GENE | CHR | P.valueA | P.valueB | cFDR.AcB | cFDR.BcA | conjunction FDR |
|---|---|---|---|---|---|---|---|---|
| rs9930506 | intronic | FTO | chr16 | 2.52E–124 | 1.90E–10 | 1.01E–123 | 1.90E–10 | 1.90E–10 |
| rs10787472 | intronic | TCF7L2 | chr10 | 3.25E–07 | 1.30E–36 | 3.25E–07 | 6.63E–35 | 3.25E–07 |
| rs2881654 | intronic | PPARG | chr3 | 1.40E–06 | 3.40E–09 | 4.19E–06 | 7.82E–08 | 4.19E–06 |
| rs849135 | intronic | JAZF1 | chr7 | 1.45E–05 | 1.70E–09 | 3.85E–05 | 7.08E–08 | 3.85E–05 |
| rs4481184 | intronic | IGF2BP2 | chr3 | 0.0002524 | 4.50E–22 | 0.0003786 | 6.55E–20 | 0.0003786 |
| rs7141420 | intronic | NRXN3 | chr14 | 8.66E–15 | 0.00025 | 4.68E–13 | 0.001125 | 0.001125 |
| rs6795735 | ncRNA_intronic | ADAMTS9–AS2 | chr3 | 2.92E–05 | 2.00E–04 | 0.000555 | 0.00604 | 0.00604 |
| rs12895330 | intergenic | AKAP6, NPAS3 | chr14 | 9.72E–05 | 0.00021 | 0.0016041 | 0.007245 | 0.007245 |
| rs2334255 | UTR3 | GIPR | chr19 | 0.0008051 | 0.00034 | 0.0114503 | 0.0176422 | 0.0176422 |
| rs1783598 | intronic | FCHSD2 | chr11 | 0.0003666 | 0.00052 | 0.0064359 | 0.0193556 | 0.0193556 |
| rs12245680 | intronic | TCF7L2 | chr10 | 0.01444 | 1.10E–09 | 0.02527 | 6.70E–07 | 0.02527 |
| rs17584208 | intergenic | PSRC1, MYBPHL | chr1 | 4.58E–06 | 0.0016 | 0.000306 | 0.02976 | 0.02976 |
| rs2488071 | intergenic | HHEX, EXOC6 | chr10 | 0.006642 | 4.70E–06 | 0.032103 | 0.0011194 | 0.032103 |
| rs11979110 | intergenic | KLF14, MIR29A | chr7 | 0.00288 | 9.70E–05 | 0.03456 | 0.0140973 | 0.03456 |
| rs1473 | intronic | PUM1 | chr1 | 0.0004889 | 0.00092 | 0.0114403 | 0.03542 | 0.03542 |
| rs10898868 | intronic | ARAP1 | chr11 | 0.002589 | 0.00044 | 0.0370227 | 0.03608 | 0.0370227 |
| rs1996023 | intergenic | GNPDA2, GABRG1 | chr4 | 1.11E–20 | 0.025 | 1.93E–17 | 0.0375 | 0.0375 |
| rs825461 | intronic | ZNF664, FAM101A | chr12 | 0.0003917 | 0.0013 | 0.0114376 | 0.04472 | 0.04472 |
| rs16945088 | intronic | FTO | chr16 | 5.30E–09 | 0.0072 | 1.48E–06 | 0.045 | 0.045 |
| rs9540493 | intergenic | LOC10272396, LINC01052 | chr13 | 3.95E–09 | 0.0057 | 1.22E–06 | 0.0456 | 0.0456 |
| rs4238585 | intergenic | GPR139, GP2 | chr16 | 1.12E–08 | 0.0069 | 3.02E–06 | 0.0483 | 0.0483 |
| rs12454712 | intronic | BCL2 | chr18 | 6.04E–06 | 0.0034 | 0.0004955 | 0.0485714 | 0.0485714 |
| rs4474658 | intergenic | C2CD4A, C2CD4B | chr15 | 0.009024 | 9.60E–06 | 0.0489874 | 0.0023849 | 0.0489874 |
Notes: P.valueA is the p value of BMI. P.valueB is the p value of T2D.
Protein-protein interaction network
The 323 identified BMI-associated genes were retrieved from the STRING database. Only 143 genes, including 46 novel genes, were annotated in this database. The 143 genes were clearly enriched in three clusters: TMEM18, PPARG and MAP2K5 (Figure S3). Two novel genes MSRA and PDILT, respectively encoding methionine sulfoxide reductase A and protein disulfide isomerase-like, were directly connected with the TMEM18 cluster. Another two novel genes, MED23 and ANPC4, respectively encoding mediator complex subunit 23 and anaphase promoting complex subunit 4, were involved in the PPARG cluster. Another three novel genes, MEF2D, RASL11A and PTPN12, respectively encoding myocyte enhancer factor 2D, RAS-like, family 11, member A and protein tyrosine phosphatase, were involved in the MAP2K5 cluster. (Figure S3).
The 89 identified T2D-associated genes were retrieved from the STRING database. Only 37 genes, including 7 novel genes, were annotated in this database. The 37 genes were clearly enriched into three clusters: HNF4A, MTNR1B and TCF7L2 (Figure S4). Three novel genes, ANXA11, BCL2L11 and NEUROG3, those respectively encoding Annexin A11, BCL2-like 11 and Neurogenin 3, were involved in the HNF4A cluster. Another two novel genes, NPBWR2 and PTHLH, encoding Neuropeptides B/W receptor 2 and Parathyroid hormone-like hormone, were involved in the MTNR1B cluster. The other novel gene MED30 was directly connected with TCF7L2 cluster (Figure S4)
Discussion
In our study, two GWASs with summary statistic p values were combined to explore the pleiotropic enrichment of SNPs that are associated with BMI and T2D. Compared to the conventional standard single phenotype GWASs, simultaneously analyzing multiple related traits allows for the increased discovery of trait-associated variants without requiring additional larger datasets for each individual trait. By leveraging the power of two different GWAS datasets from BMI and T2D, we discovered 287 loci for BMI and 75 loci for T2D. Using the standard GWAS significance threshold in the datasets, only 3631 were significant for BMI. Most of the genes have not been reported to show borderline significance with BMI, as detailed in Table S2. Adopting the genetic pleiotropy-informed cFDR method, we found 12 additional novel loci associated with both BMI and T2D. These novel findings may enable us to further dissect the overlapping genetic mechanisms between these two related phenotypes. The improved detection of novel susceptibility loci with genetic pleiotropy may lead us to a better understanding of common etiology between disorders and have a significant impact on the clinical treatment and prevention of related complex human diseases.
The cFDR approach was adopted here to account for some of the missing heritability between traits or diseases. This method employs the idea that a variant with significant effects in two associated phenotypes is more likely to be a true effect, and therefore has a higher probability of being detected in multiple independent studies4,7. This technique allows for an increase in effective sample size and therefore a subsequent increase in power to detect true associations for more variants with small to moderate effect sizes, which are often ignored in the standard single phenotype GWAS. In addition, the genetic enrichment presented in conditional Q-Q plots conveys that the decreased cFDR value for a given nominal p value greatly increases power to detect true association effects. When initially implementing the cFDR method, Andreassen et al.7 demonstrated one advantage of this model-free empirical cdf approach is for the avoidance of bias in cFDR estimates from model misspecification. Through a comparison of traditional unconditional FDR and cFDR methods, they found that the latter resulted in an increase of 15–20 times the number of SNPs under the same FDR threshold of 0.057.
Our cFDR analysis identified 23 pleiotropic signals annotated to 30 genes, providing evidence for the close relationship and shared genetic mechanisms between these two traits. These findings are consistent with the evidence from previous studies25 that have demonstrated a causal relationship between these two traits. The genes FTO, MC4R and TCF7L2 were frequently reported and replicated in previous BMI and T2D related studies26,30,41. However, potential confounding factors and biases might coincidently be responsible for some of these associations. For the genes FTO and MC4R, their respective effects on T2D were found to be modest and previous studies showed that their effects on T2D disappeared after adjustment for BMI42. In European populations, TCF7L2 was not reported as a risk factor for obesity although its effect on T2D risk is modulated by obesity because of the interaction between TCF7L2 polymorphisms (rs7903146) and BMI status43. The implementation of the cFDR method in our study not only furnishes another empirical validation for the cFDR method to successfully detect novel and known disease associated genetic variants, but also shows the possibility of improved discovery of novel susceptibility loci using existing GWAS summary statistics. There were 14 genes (JAZF1, IGF2BP2, NRXN3, ADAMTS9-AS2, GIPR, FCHSD2, HHEX, EXOC6, KLF14, ARAP1, GNPDA2, GP2, C2CD4A and C2CD4B) that were associated with either BMI or T2D in previous studies but not with both that were detected as pleiotropic loci in this analysis. Furthermore, 12 novel genes are worth noting because no previous study has reported associations with either BMI or T2D for any of them. For the SNPs that were annotated to these 12 genes, two SNPs were located in the intronic regions of genes ZNF664 and PUM1 and the other five SNPs were all located in intergenic regions. As examples, we will discuss two of these genes ZNF664 and PUM1 for their potential functional relevance and significance.
The SNP rs825461 is located at the intronic region between gene ZNF664 and FAM101A. The ZNF664 gene encodes a protein named zinc finger protein 664, and one study reported that ZNF664 was involved in eye development and that the monogenic form may be associated with high risk of myopia44. Furthermore, ZNF664 was previously reported to show suggestive association (P < 1E-4) with adiponectin45, a protein involved in many metabolic processes including glucose regulation and fatty acid oxidation46. The rs1473 SNP is located at the intronic region of the gene PUM1, a member of the PUF family of proteins that contains a sequence-specific RNA binding domain. One study reported that the protein may be involved in the regulation of embryogenesis, and cell development and differentiation47. These genes may be involved in certain processes that are significant in the development of obesity and T2D, however future studies are needed to explore the exact mechanisms of those novel genes we identified.
Our study presents several strengths. First, the statistical power is increased through the cFDR method by leveraging two large GWAS datasets, providing an increase in the effective sample size. Although a meta-analysis of the same data would offer a similar gain, the meta-analysis approach only allows for more powerful detection of loci with the same direction of allelic effects in the phenotypes48, whereas the cFDR method allows for detection of loci regardless of their effect directions. Second, we consider two traits that are unlikely to be correlated with BMI and T2D, ADHD and MDD, and generate conditional QQ plots with respect to these “control traits.” The “control trait” enrichment analysis provides an alternative way to examine pleiotropic enrichment and provides a baseline that can be used to statistically partially validate the novel findings in our study. We believe that the collider-stratification bias is unlikely in our analysis because, the GWAS datasets have undergone genomic control (GC) and we also carried out LD pruning with r2 > 0.2. In addition, our conditional analysis provides a model-free method to obtain conservative estimation4,7,8.
Our study may also have some important limitations. First, we could not provide information about the effect estimates of pleiotropic loci on the phenotypes due to a lack of detailed individual-study-level data. However, we can infer this information from the summary effect sizes in the original GWAS study. This cFDR approach cannot distinguish between vertical and horizontal pleiotropy of the pleiotropic signals, although this question might be partially addressed in future Mendelian Randomization49,50 studies. Second, it is likely that some of our cFDR results may be overestimated due to overlapping samples although the model-free approach is able to neutralize this overestimation of the conservative cFDR estimate4,7,8. Alternative approaches may be applied to check whether novel loci could still be identified in order to further confirm novel findings in our study or to furnish an empirical comparison of the relative performance of alternative methods, a topic we wish to pursue in the future with comprehensive theoretical and simulation approaches.
In summary, by incorporating the shared genetic effects of two closely related traits into a conditional analysis framework, we observed significant pleiotropic enrichment between obesity and T2D. We identified several novel pleiotropic loci of potential functional significance for obesity and T2D in our analysis, and the results may provide us with novel insights into the shared genetic influences between these two disorders.
Materials and Methods
GWAS Datasets
The dataset for T2D contains association summary statistics for a trans-ethnic T2D GWAS meta-analysis of 26,488 cases and 83,964 controls30. Ancestry-specific meta-analyses were previously performed by component datasets from the full set of cohorts, including the DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium (European descent)29, the Asian Genetic Epidemiology Network T2D (AGEN-T2D) Consortium (East Asian descent)51, the South Asian T2D (SAT2D) Consortium (south Asian descent)52, and the Mexican American T2D (MAT2D) Consortium (Mexican and Mexican American descent)53. Further details of the samples and methods employed within each ancestry group are presented in the corresponding consortium papers29,51–53. Briefly, various genotyping products were applied in the individual study’s assay processes, with appropriate sample and SNP quality control (QC). Genotype imputation was conducted within each GWAS dataset using Phase II/III HapMap as the reference panels. Each SNP with MAF > 1% (except MAF > 5% in MAT2D GWAS due to a smaller sample size) and passing QC was analyzed for association with T2D using an adjusted additive model. Association summary statistics of each ancestry-specific meta-analysis were combined using a fixed-effect inverse-variance weighted meta-analysis. Genomic control (GC) was carried at the individual study level, after ethnicity-specific meta-analysis, and finally after trans-ethnic meta-analysis30.
The GIANT dataset for BMI contains association summary statistics for the GWAS and Metabochips meta-analysis of 339,224 individuals of various ancestries, including 322,154 individuals of European descent and 17,072 individuals of African-American and Hispanic descent31. The data contains the summary p-values from meta-analysis after correction for inflation due to potential population admixture. Two rounds of GC were separately applied both at the cohort level and after meta-analysis31.
Data Preparation
Before the implementation of the cFDR method, several preparation steps were performed. First, we checked the European Ancestry cohorts for overlapping samples included in these two datasets (Table S1). Next, we combined the common SNPs included in these two datasets. Then we applied a linkage disequilibrium (LD) based SNP pruning method7,8 to remove large correlations between pairs of variants. The SNP pruning method begins using a window of 50 SNPs where LD is calculated between each pair of SNPs. The minor allele frequency (MAF) is the basis for the SNP pruning, where for pairs with r2 > 0.2 we removed the SNP with smaller MAF. Following this initial removal of SNPs, the window slides 5 SNPs forward and the process is repeated until there are no pairs of SNPs that are high in LD. The dataset was pruned using the HapMap 3 genotypes as a reference panel. Last, we performed gene annotation for the final set of 123,804 variants that were included in the analyses.
GC corrections were used in the GWASs to ensure that the variance estimates for each SNP are not inflated due to population structure and cryptic relatedness54. Both of the original datasets30,31 we adopted in our study applied GC at the individual study level and again after meta-analysis, hence there was no need for us to reapply the GC in this analysis.
Statistical analysis
The cFDR approach is well-established now, which has been widely applied by many other groups4,7,8,55,56 and our group12–14. We briefly summarized this cFDR approach as follows: after the data preparation processing, we computed the conditional empirical cumulative distribution functions (cdfs) of the corrected p-values for the x axis in conditional QQ plot. Empirical cdfs for BMI SNP p-values were conditioned on nominal p-values in T2D, and vice versa. For each nominal p-value, an estimate of the cFDR was obtained from the conditional empirical cdfs. Using this cFDR approach, we obtained two cFDR tables–cFDR result for BMI conditioned on T2D and vice versa. Using these tables we identified loci associated with BMI and T2D (cFDR < 0.05), respectively. Then a conjunction method was used to find SNPs significantly associated with both BMI and T2D. Specifically, we took the maximum of those two cFDR values above as our conjunction FDR.
Conditional QQ and enrichment plots for assessing pleiotropic enrichment
As an intuitive illustration, we presented conditional Q-Q plots to graphically assess the pleiotropic enrichment of SNPs of the principal phenotype successively conditioning on various strengths of associations with the conditional phenotype. We plotted the QQ curve for the quantiles of nominal -values obtained from GWAS summary statistics for association of the subset of SNPs that are below each significance threshold in the conditional trait. The nominal -values were plotted on the y-axis and the empirical quantiles (empirical cumulative distribution functions (cdfs)) of the nominal p-values were plotted on the x-axis. Under the global null hypothesis, the theoretical distribution of p-values is expected to lie approximately on the diagonal line of the Q-Q plots. Enrichment of genetic associations is indicated as a leftward deflection from the null line as the principal phenotype is successively conditioned on increasing strength of associations with the conditional phenotype. The degree of deflection between curves provides important information about the degree of pleiotropy between the two phenotypes. Larger deflections are considered to represent a greater enrichment of pleiotropic genes between the two phenotypes.
For the associated phenotypes BMI and T2D, pleiotropic “enrichment” of BMI with T2D exists if the proportion of SNPs or genes associated with BMI increases as a function of increased association with T2D. To confirm the pleiotropic enrichment effect, we presented fold-enrichment plots of nominal values for BMI SNPs below the standard GWAS threshold of p < 5 × 10−8 and for subsets of SNPs determined by the significance of their association with T2D and vice versa. As the p values of the conditional phenotypes become more significant, lower upward shift from the null line will persist.
In order to check the pleiotropic enrichment and provide a baseline that can be used to confirm novel findings, we also generated conditional QQ plots for two control traits that are unlikely to be correlated with BMI and T2D, ADHD and MDD.
Conditional Manhattan plots for localizing genetic variants
To demonstrate the localization of the SNPs associated with BMI conditional on their significance on T2D, and the reverse, we present conditional Manhattan plots. The plots present the relationship between all SNPs within an LD block and their chromosomal locations. The 22 chromosomal locations are plotted on the x-axis, and the BMI values conditional on T2D are plotted on the y-axis and vice versa for T2D. Any SNP with a value greater than 1.3 (FDR < 0.05) was deemed to be significantly associated with the principal phenotype. We also present a conjunction Manhattan plot to demonstrate the locations of the common pleiotropic genetic variants associated with both phenotypes.
Functional annotation and gene enrichment analysis
In order to evaluate the biological functions of the individual trait associated loci identified by cFDR and pleiotropic loci identified by conjunction FDR, we performed functional annotation and gene enrichment analysis using the gene ontology (GO) terms database (http://geneontology.org/)57. All significant genes identified by cFDR and conjunction cFDR in our study were annotated and characterized based on three main categories: biological processes, cellular component and molecular functions. This analysis provided comprehensive biological information, allowing us to partially validate our findings by determining specific genes that are enriched in T2D- and obesity-related GO terms.
Protein-protein interaction network
In order to detect interactions and associations of the BMI-associated and T2D-associated genes respectively, protein-protein interaction analyses were conducted by searching the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/). The STRING database comprises known and predicted associations from curated databases or high-throughput experiments, and also with other associations derived from text mining, co-expression, and protein homology58.
Electronic supplementary material
Acknowledgements
This research was supported by the Study on Emergency Compensation Mechanism of Major Public Health Epidemic (Grant NO: 152102310049). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Meanwhile, the authors would like to thank the participants, the coordinators, and administrators for their supports during the study.
Author Contributions
Q.Z. as the first author performed data analysis and wrote the manuscript. K.H.W., J.Y.H., X.X., X.L., H.M.L., and W.Q.L. provided advice and suggestions while we met some problems during the data analysis process, and Y.Z. and J.G. contributed suggestions for manuscript revision and revised the manuscript. W.D.Z., Y.L.X., X.Z.S., C.Q.S. all gave constructive suggestions during the whole process. C.Q.S. and H.W.D. are the co-corresponding authors. H.W.D. conceived and initiated this project, provided advice on experimental design, oversaw the implementation of the statistical method, and revised/finalized the manuscript. C.Q.S. help revised the manuscript and oversaw the point to point response to reviewers in our rebuttal later.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16722-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chang-Qing Sun, Email: suncq@zzu.edu.cn.
Hong-Wen Deng, Email: hdeng2@tulane.edu.
References
- 1.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoo YJ, Pinnaduwage D, Waggott D, Bull SB, Sun L. Genome-wide association analyses of North American Rheumatoid Arthritis Consortium and Framingham Heart Study data utilizing genome-wide linkage results. BMC proceedings. 2009;3(Suppl 7):S103. doi: 10.1186/1753-6561-3-s7-s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreassen OA, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cichonska, A. et al. metaCCA: Summary statistics-based multivariate meta-analysis of genome-wide association studies using canonical correlation analysis bioRxiv, 10.1101/022665 (2015). [DOI] [PMC free article] [PubMed]
- 6.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. doi: 10.1371/journal.pgen.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreassen OA, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreassen OA, et al. Identifying common genetic variants in blood pressure due to polygenic pleiotropy with associated phenotypes. Hypertension. 2014;63:819–826. doi: 10.1161/HYPERTENSIONAHA.113.02077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stearns FW. One hundred years of pleiotropy: a retrospective. Genetics. 2010;186:767–773. doi: 10.1534/genetics.110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium C-DGotPG. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng, C. et al. Enhanced Identification of Potential Pleiotropic Genetic Variants for Bone Mineral Density and Breast Cancer. Calcified tissue international, 10.1007/s00223-017-0308-x (2017). [DOI] [PMC free article] [PubMed]
- 12.Peng C, et al. Genetic sharing with coronary artery disease identifies potential novel loci for bone mineral density. Bone. 2017;103:70–77. doi: 10.1016/j.bone.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenbaum J, et al. Increased detection of genetic loci associated with risk predictors of osteoporotic fracture using a pleiotropic cFDR method. Bone. 2017;99:62–68. doi: 10.1016/j.bone.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng, C. P. et al. Increased identification of novel variants in type 2 diabetes, birth weight and their pleiotropic loci. Journal of diabetes, 10.1111/1753-0407.12510 (2016). [DOI] [PMC free article] [PubMed]
- 15.Taylor AE, et al. Comparison of the associations of body mass index and measures of central adiposity and fat mass with coronary heart disease, diabetes, and all-cause mortality: a study using data from 4 UK cohorts. The American journal of clinical nutrition. 2010;91:547–556. doi: 10.3945/ajcn.2009.28757. [DOI] [PubMed] [Google Scholar]
- 16.Ng M, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus–present and future perspectives. Nature reviews. Endocrinology. 2012;8:228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- 18.Golay A, Ybarra J. Link between obesity and type 2 diabetes. Best Pract Res Clin Endocrinol Metab. 2005;19:649–663. doi: 10.1016/j.beem.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almgren P, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54:2811–2819. doi: 10.1007/s00125-011-2267-5. [DOI] [PubMed] [Google Scholar]
- 21.Karaderi T, Drong AW, Lindgren CM. Insights into the Genetic Susceptibility to Type 2 Diabetes from Genome-Wide Association Studies of Obesity-Related Traits. Curr Diab Rep. 2015;15:83. doi: 10.1007/s11892-015-0648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-Indias I, Cardona F, Tinahones FJ, Queipo-Ortuno MI. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Frontiers in microbiology. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 24.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 25.Corbin LJ, et al. BMI as a Modifiable Risk Factor for Type 2 Diabetes: Refining and Understanding Causal Estimates Using Mendelian Randomization. Diabetes. 2016;65:3002–3007. doi: 10.2337/db16-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamura M, et al. Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat Commun. 2016;7:10531. doi: 10.1038/ncomms10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer F, et al. Obesity genes identified in genome-wide association studies are associated with adiposity measures and potentially with nutrient-specific food preference. The American journal of clinical nutrition. 2009;90:951–959. doi: 10.3945/ajcn.2009.27781. [DOI] [PubMed] [Google Scholar]
- 29.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–U401. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall JC, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nature Genetics. 2013;45:1274-+. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaulton, K. J. et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. 47, 1415–1425, 10.1038/ng.3437 (2015). [DOI] [PMC free article] [PubMed]
- 36.Scott RA, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–U378. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxena R, et al. Large-Scale Gene-Centric Meta-Analysis across 39 Studies Identifies Type 2 Diabetes Loci (vol 90, pg 410, 2012) American Journal of Human Genetics. 2012;90:753–753. doi: 10.1016/j.ajhg.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strawbridge RJ, et al. Genome-Wide Association Identifies Nine Common Variants Associated With Fasting Proinsulin Levels and Provides New Insights Into the Pathophysiology of Type 2 Diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant SF, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 42.Cauchi S, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. Journal of molecular medicine. 2009;87:537–546. doi: 10.1007/s00109-009-0451-6. [DOI] [PubMed] [Google Scholar]
- 43.Cauchi S, et al. Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity. 2008;16:476–482. doi: 10.1038/oby.2007.77. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, et al. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 2011;7:e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y, et al. A meta-analysis of genome-wide association studies for adiponectin levels in East Asians identifies a novel locus near WDR11-FGFR2. Human Molecular Genetics. 2014;23:1108–1119. doi: 10.1093/hmg/ddt488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diez JJ, Iglesias P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur J Endocrinol. 2003;148:293–300. doi: 10.1530/eje.0.1480293. [DOI] [PubMed] [Google Scholar]
- 47.Parisi M, Lin H. The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics. 1999;153:235–250. doi: 10.1093/genetics/153.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawlor DA. Commentary: Two-sample Mendelian randomization: opportunities and challenges. International Journal of Epidemiology. 2016;45:908–915. doi: 10.1093/ije/dyw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kemp JP, Sayers A, Smith GD, Tobias JH, Evans DM. Using Mendelian randomization to investigate a possible causal relationship between adiposity and increased bone mineral density at different skeletal sites in children. Int J Epidemiol. 2016;45:1560–1572. doi: 10.1093/ije/dyw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho YS, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kooner JS, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parra EJ, et al. Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican-American sample from Starr County, Texas. Diabetologia. 2011;54:2038–2046. doi: 10.1007/s00125-011-2172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 55.Le Hellard S, et al. Identification of Gene Loci That Overlap Between Schizophrenia and Educational Attainment. Schizophrenia bulletin. 2017;43:654–664. doi: 10.1093/schbul/sbw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smeland, O. B. et al. Identification of genetic loci shared between schizophrenia and the Big Five personality traits. 7, 2222, 10.1038/s41598-017-02346-3 (2017). [DOI] [PMC free article] [PubMed]
- 57.Consortium GO. Gene Ontology Consortium: going forward. Nucleic acids research. 2015;43:D1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franceschini A, et al. STRINGv9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.