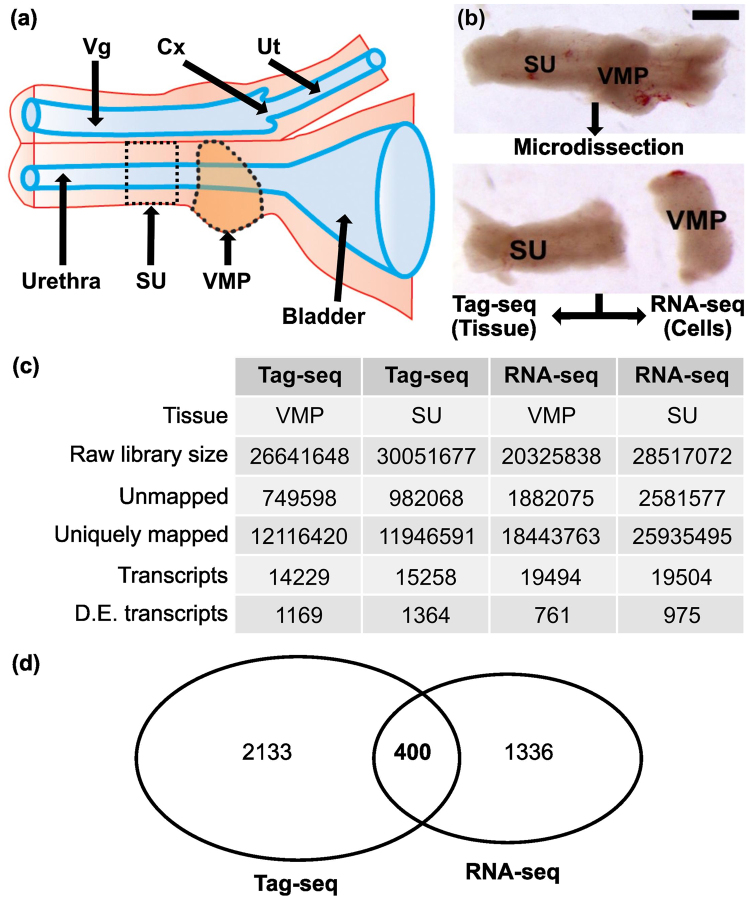
Figure 1.
Identification of transcripts differentially expressed in VMP and SU tissues using Tag-seq and RNA-seq. (a) A schematic diagram of P0 female urethra illustrating the position of the VMP on the urethra, and an adjacent region of urethra comprised of smooth muscle and urethral epithelia (SU). Vg = Vagina, Cx = Cervix, Ut = Uterus; only one uterine horn is illustrated. (b) P0 urethra was micro-dissected to yield VMP mesenchyme and adjacent SU. Micro-dissected VMP consists of pure mesenchyme and lacks epithelia, while SU contains both mesenchyme and epithelia. Tag-seq libraries were prepared from microdissected VMP and SU tissues (Tissue), while RNAseq libraries were prepared from cell suspensions from tissue digested with collagenase (Cells). This enabled the removal of urethral epithelium from the SU cell sample. (c) Details of Tag-seq and RNA-seq libraries, and differentially expressed transcripts. A statistical package, NOISeq, was used to define differentially expressed (DE) transcripts in both Tag-seq and RNA-seq datasets. Transcripts with an absolute log2 fold-change (M) ≥ 1.5 and a diverge probability (q) > 0.9 were considered to be differentially expressed. (d) Venn diagram showing overlap between DE transcripts in Tag-seq vs RNA-seq data, and identification of 400 DE transcripts common to both techniques.