Abstract
Aim:
We tested whether genetic variation in miRNA processing genes modified the association of PM2.5 with DNA methylation (DNAm) age.
Patients & methods:
We conducted a repeated measures study based on 552 participants from the Normative Aging Study with multiple visits between 2000 and 2011 (n = 940 visits). Address-level 1-year PM2.5 exposures were estimated using the GEOS-chem model. DNAm-age and a panel of 14 SNPs in miRNA processing genes were measured from participant blood samples.
Results & conclusion:
In fully adjusted linear mixed-effects models, having at least one copy of the minor rs4961280 [AGO2] allele was associated with a lower DNAm-age (β = -1.13; 95% CI: -2.26 to -0.002). However, the association of PM2.5 with DNAm-age was significantly (Pinteraction = 0.01) weaker in homozygous carriers of the major rs4961280 [AGO2] allele (β = 0.38; 95% CI: -0.20 to 0.96) when compared with all other participants (β = 1.58; 95% CI: 0.76 to 2.39). Our results suggest that miRNA processing impacts DNAm-age relationships.
Keywords: : AGO2, air pollution, epigenetic aging, miRNA, PM2.5, SNPs
Graphical abstract:
miRNA processing AGO2 polymorphism (rs4961280) modifies the association of long-term ambient fine particle exposure with blood DNA methylation age
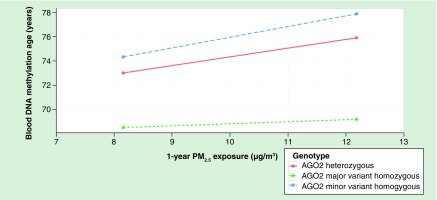
The graph depicts lines from a fully adjusted linear regression model with fine particle exposure levels ranging from the tenth to the ninetieth percentile, all other continuous variables held constant at their means, and all other categorical variables held at their most frequent level.
Between 2015 and 2050, the global percentage of individuals over the age of 60 is expected to almost double from 12 to 22% [1]. This shift in the age composition of the global population is particularly important because it will likely be accompanied by immense public health and economic burdens due to an unprecedented volume of aging-related diseases. For instance, in the USA alone, the 2017 disease prevalence and economic costs associated with Alzheimer's dementia were 5.5 million and $259 billion. By 2050, those statistics are expected to be 16 million and $1.1 trillion, respectively [2]. In an effort to curb these expanding disease and economic burdens, there has been an ever growing emphasis on research aimed at understanding biological aging and the factors that contribute to adverse aging-related health outcomes.
Ambient fine particle air pollution (PM2.5) – often considered the world's largest singular environmental health risk – is one potentially modifiable risk factor for aging-related diseases including cardiovascular disease, cognitive decline and cancer [3–6]. Of particular interest, is the association of long-term PM2.5 exposure levels with DNA methylation (DNAm) age, a novel epigenome-wide DNA methylation-based measure of biological aging [7,8]. Like other biomarkers of biological aging, DNAm-age has been associated with all-cause mortality and aging-related diseases [9,10]. In contrast to other biomarkers of aging, researchers remain highly uncertain about what DNAm-age is capturing on a molecular physiological level [11]. By examining the relationship of DNAm-age with PM2.5, a widely studied exogenous exposure and aging risk factor, we can begin to understand more about DNAm-age physiology.
As previously mentioned, DNAm-age is derived from measurements of DNA methylation. DNA methylation is a biological process where methyl groups are added to DNA nucleotides and often result in changes in gene expression [12]. miRNAs are small noncoding RNA molecules that can also regulate gene expression and have been associated with PM2.5, aging/aging-related diseases and DNA methylation [13–15]. miRNAs are produced from nuclear transcripts that form hairpin structures. Following nuclear and cytoplasmic processing by a series of enzymes, miRNAs are incorporated into a structure called the RNA-induced silencing complex (RISC). RISC achieves post-transcriptional gene regulation by using one strand of the incorporated miRNA to target mRNAs via nucleotide complementary base pairing. Once the relevant mRNA is targeted, RISC inhibits subsequent protein production by inducing mRNA cleavage or by reducing translation of the mRNA molecule [16].
Despite existing knowledge of the role of miRNAs in PM2.5 and aging biology, studies have not yet examined the role of miRNAs in the PM2.5–DNAm-age relationship. Given the shared role of miRNAs and DNA methylation in gene regulation [17], we hypothesized that miRNA physiology would be related to DNAm-age and may play a role in the relationship of PM2.5 with DNAm-age. In the present study, we investigated if SNPs in miRNA processing genes modified the associations of long-term PM2.5 and PM2.5 component species (sulfate and ammonium) exposure with DNAm-age in participants of the elderly Normative Aging Study (NAS). Rather than simply testing all the SNPs in our panel, we utilized a methodical framework to identify and analyze significant SNPs and PM2.5–SNP interactions. We first employed an elastic net (penalized regression) selection model to identify SNPs in miRNA processing genes that were specifically important to DNAm-age. Subsequently, we used fully adjusted linear mixed-effects models to test for statistically significant direct associations of the elastic net selected SNPs and PM2.5–SNP interactions with DNAm-age. We also conducted a number of secondary analyses to better ascertain if particular PM2.5 component species were responsible for the relationships we observed.
Methods
Study population
The US Department of Veterans Affairs (VA) NAS is a longitudinal study of aging that was established in 1963 and recruited male participants from the Greater Boston area that were free of any chronic disease [18]. The NAS is now a closed cohort, but every 3–5 years since recruitment, participants return for onsite, follow-up study visits. During these recurring visits, participants undergo thorough physical examinations, report lifestyle practices via questionnaires, and provide biospecimens including blood. At recruitment, all participants provided written informed consent to the VA Institutional Review Board and were at least 18 years of age. The VA and Harvard T.H. Chan School of Public Health Institutional Review Boards granted human subjects approval (protocol 14027–102).
Our study sample is derived from all NAS men with continued study participation since the year 2000, when address-level PM2.5 component species estimates became available. We started with a total of 552 participants with 940 study visits (observations) between the years 2000 and 2011 [8]. Of these 552 participants, 249 (45%) had one study visit, 218 (40%) had two study visits and 85 (15%) had three or more study visits. From this sample, we then excluded participants missing miRNA processing gene polymorphism data. This resulted in a final study sample of 471 participants with 808 total study visits. In the final study sample, 208 participants (44%) had one visit, 189 (40%) had two visits and 74 (16%) had three or more visits.
Measuring DNA methylation & computing DNAm-age
We extracted DNA from whole blood provided by participants during NAS visits. After performing bisulfite conversion on the DNA (EZ-96 DNA Methylation Kit, Zymo Research, CA, USA), we performed methylation analysis using the Illumina HumanMethylation450 BeadChip platform (Infinium HD Methylation protocol, Illumina, CA, USA). To ensure a similar age distribution across chips/plates and minimize batch effects, we used a two-stage age-stratified algorithm to randomize samples and randomized chips across plates. For quality control purposes, we removed samples where more than 5% of probes had a beadcount less than 3 or more than 1% of probes had a detection p-value > 0.05. The remaining samples were preprocessed with Illumina-type background correction without normalization and normalized with dye-bias and BMIQ3 adjustments. Next, we generated methylation β-values, which represent the percentage of methylation for each of the approximately 480,000 CpG sites in the BeadChip array. β = intensity of the methylated signal (M)/[intensity of the unmethylated signal (U) + intensity of the methylated signal (M) + 100].
DNAm-age was computed using the publicly available online calculator [19]. DNAm-age was derived from a penalized regression (an elastic net) run on numerous datasets of diverse cell and tissue types where CpG probes shared by both the Illumina HumanMethylation27 and HumanMethylation450 BeadChip platforms were regressed on a calibrated version of chronological age. Three hundred and fifty-three CpGs that correlated with age (193 positively and 160 negatively) were selected by the elastic net [11]. The model coefficients from these 353 CpGs were used by the calculator to predict the age of each DNA sample (i.e., DNAm-age). The calculator maintains predictive accuracy (age correlation 0.97, error = 3.6 years) across almost all body tissues including blood, bone, and brain [11].
One-year address-level ambient fine particulate matter (PM2.5) exposure estimation
We focused on the 1-year PM2.5 exposure window because existing literature demonstrates that it is robustly associated with DNA-age [7]. Furthermore, >90% of NAS participants are retired; thus, home address exposures are expected to be a good proxy for their individual ambient exposures. Using the GEOS-chem chemical transport model [20], we generated daily estimates at the 1 km × 1 km area resolution for total PM2.5. The GEOS-chem model is particularly useful because it allows us to predict PM2.5 component species like ammonium and sulfate at the same 1 km × 1 km area resolution. Sulfate and ammonium are the major PM2.5 component species that have been previously shown to be important in predicting DNAm-age [8]. After geocoding and linking participants’ residencies to an area-level grid point, we assigned particle estimates to each participant's address. One-year total PM2.5 and PM2.5 component species exposure estimates were determined by averaging daily exposures for the 365 days prior to the day of each participants’ NAS visit. Tenfold cross-validation demonstrated that the model performed well for PM2.5 mass and its component species with R2s ranging from 0.70 to 0.88 [21].
Genotyping miRNA processing gene polymorphisms
The panel of 24 miRNA processing gene SNPs examined in this study were selected from previous studies that investigated the association of miRNA processing gene SNPs with chronic aging-related diseases [22,23]. Some of these same SNPs have been shown to modify relationships of ambient air pollutants with aging-related disease [24,25]. We performed genotyping on DNA extracted from participants’ blood. Multiplex PCR assays were designed with Sequenom SpectroDESIGNER software (Sequenom, Inc., CA, USA). The extension product was subsequently spotted onto a 384-well spectroCHIP and analyzed in the MALDI-TOF mass spectrometer (Sequenom, Inc.). We duplicated the assay for 5% of the samples. All 24 SNPs analyzed for this study were successfully detected.
Following genotyping, we excluded ten SNPs for which the number of participants who were homozygous minor variant carriers was less than ten (rs595961 and rs636832 in AGO1; rs197388 and rs197414 in DDX20; rs417309 in DGCR8; rs3742330 in DICER1; rs2740348 and rs3744741 in GEMIN4; rs1106042 in PIWIL1) and one in which Hardy–Weinberg equilibrium was not met at the 0.05 level (rs10719 in DROSHA). This exclusion criterion has been utilized in already published studies that use this panel of SNPs [24,25]. The remaining 14 SNPs were used in the study analyses. Linkage disequilibrium of SNPs within the same gene was previously assessed using the LDPlotter tool [26] [25].
Statistical analysis
Elastic net selection of miRNA processing gene SNPs
The aim of the present study was to examine if SNPs in miRNA processing genes modified the association of long-term PM2.5 and PM2.5 component species levels with DNAm-age. In an effort to limit multiple comparisons; and identify specific miRNA processing gene SNPs that are important to DNAm-age, we first employed an elastic net (penalized regression) via the glmnet function in the R glmnet package. Our elastic net method was similar to that described by Lenters and colleagues [27] and the full documentation for running all aspects of the elastic net via glmnet is publicly available [28]. We have also used a comparable elastic net strategy in a previous publication [29]. In short, the elastic net regression linear models utilized a hybrid of ridge and LASSO penalty functions to determine which SNPs were important to DNAm-age. By combining both of these penalty functions, the elastic net is able to perform selection while allowing for the inclusion of highly related genetic variants [30,31]. In our case, the highly related variants were the panel of miRNA processing gene SNPs. Specifically, in our elastic net selection model, all 14 SNPs and their respective PM2.5–SNP interactions were regressed on DNAm-age. Chronological age (continuous), blood cell proportions (plasma cells, CD4+ lymphocytes, CD8+ lymphocytes, natural killer cells, monocytes and granulocytes) (continuous, determined via Houseman and Horvath methods [11,32]), average 1-year temperature (continuous address-specific satellite measurements [8]), cumulative cigarette pack years (continuous), smoking status (current, former or never), and season of visit (spring [March–May], Summer [June–August], Fall [September–November] and Winter [December–February]), were also included in the selection model as unpenalized variables. The existing air pollution, DNA methylation and DNAm-age literature have identified these variables as important potential confounders [7–8,33–34]. Cross-validation was performed to determine the optimal degree of penalization and the minimum mean-squared error of prediction from repeated tenfold cross-validation was used in the final elastic net selection model. miRNA processing gene SNPs with nonzero model coefficients were considered as ‘selected’ by the elastic net.
Covariates
The direct relationships of the miRNA processing gene SNPs with DNAm-age and the role of these SNPs as modifiers of the association of PM2.5 with DNAm-age were examined using fully adjusted linear mixed-effects models. These models included a random participant-specific intercept to account for correlation between repeated outcome measures resulting from having multiple study visits for participants. In the analyses using fully adjusted models, we controlled for all the variables used in the elastic net selection model as well as BMI (lean [<25], overweight [25–30], obese [>30]), alcohol intake (yes/no more than or equal to two drinks daily), maximum years of education (continuous), cancer (yes/no history of lifetime cancer diagnosis), ischemic heart disease (yes/no based on electrocardiogram, validated medical records or physical exam), diabetes (physician diagnosis or a fasting blood glucose >126 mg/dl) and hypertension (yes/no antihypertensive medication use or systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg).
Effect modification by & direct associations of miRNA processing gene SNPs
Using the same covariates from the direct effect fully adjusted linear mixed-effects models, we evaluated if the miRNA processing gene SNPs selected by the elastic net modified the association of PM2.5 with DNAm-age. Given a statistically significant modifying effect, we evaluated if the SNP of interest modified the associations of sulfate and ammonium with DNAm-age. In these analyses the reference group was participants who were homozygous for the major variant of the SNP. For instance, if we evaluated the modifying role of ‘SNP A’ on the association of PM2.5 with DNAm-age, we compared participants genotyped as homozygous for the major variant of ‘SNP A’ against all other participants (i.e., participants genotyped heterozygous or homozygous for the minor variant of ‘SNP A’). We conducted secondary analyses exploring trends in significant modifier effects across all three genotypes (homozygous major variant, heterozygous and homozygous minor variant). When we observed that a miRNA processing gene SNP significantly modified the relationship of a PM2.5 component species with DNAm-age, we also conducted an additional sensitivity analysis. In this sensitivity analysis, we subtracted that component species from total PM2.5 and re-evaluated effect modification by the SNP.
Also using fully adjusted linear mixed-effects models, we determined if elastic net-selected miRNA processing gene SNPs had direct associations with DNAm-age when modeled as joint predictors with PM2.5 levels. We also performed a sensitivity analysis, examining these direct associations, where we subtracted component species from total PM2.5 as previously described above.
Network analysis
We used the publicly available GeneMANIA platform [35] to explore gene network relationships (co-expression, physical interactions and genetic interactions) between the genes encompassing the elastic net-selected SNPs and the 353 genes that contribute CpGs to the DNAm-age metric.
Analysis software
The gene network analysis was performed using the Genemania plugin for Cytoscape. All remaining statistical analyses were performed using R Version 3.1.1 (R Core Team, Vienna, Austria) and we considered a p-value < 0.05 to be statistically significant.
Results
Descriptive statistics
Table 1 summarizes the demographic and clinical characteristics of study participants across all study visits. All participants were Caucasian males with a mean (standard deviation [SD]) chronological age and DNAm-age of 75.0 (7.03) and 74.1 (8.02). A majority of the participants completed at least 12 years of formal education (74%), were former smokers (67%) and were overweight or obese (77%). In this study sample, the prevalences of ischemic heart disease, diabetes and hypertension were 35, 18 and 75%, respectively. The lifetime prevalence of a cancer diagnosis was 57%.
Table 1. . Characteristics of study participants (2000–2011).
| Main variables | All visits, N = 808 |
|---|---|
| Age (years), mean (SD) | 75.0 (7.03) |
| DNAm-age (years), mean (SD) | 74.1 (8.02) |
| 1-year fine particle level (μg/m3), mean (IQR): | |
| – PM2.5 | 10.3 (2.15) |
| – Sulfate | 3.39 (0.81) |
| – Ammonium | 1.05 (0.29) |
| Lifestyle and environmental variables | |
| Alcohol consumption, N (%): | |
| – <2 drinks/day | 647 (80) |
| – ≥2 drinks/day | 161 (20) |
| BMI, N (%): | |
| – Healthy/lean | 189 (23) |
| – Overweight | 427 (53) |
| – Obese | 192 (24) |
| Education, N (%): | |
| – ≤12 years | 206 (26) |
| 12–16 years | 379 (47) |
| – >16 years | 223 (27) |
| Pack years, mean (SD) | 21.2 (24.7) |
| Smoking status, N (%): | |
| – Current | 36 (4) |
| – Former | 538 (67) |
| – Never | 234 (29) |
| Season, N (%): | |
| – Spring | 204 (25) |
| – Summer | 175 (22) |
| – Fall | 271 (33) |
| – Winter | 158 (20) |
| Temperature (°C), mean (SD) | 11.3 (0.99) |
| Aging-related diseases | |
| Ischemic heart disease, N (%): | |
| – Yes | 287 (35) |
| – No | 521 (65) |
| Diabetes, N (%): | |
| – Yes | 148 (18) |
| – No | 660 (82) |
| Hypertension, N (%): | |
| – Yes | 604 (75) |
| – No | 204 (25) |
| Lifetime cancer diagnosis, N (%): | |
| – Yes | 458 (57) |
| – No | 350 (43) |
DNAm: DNA methylation; IQR: Interquartile range; SD: Standard deviation.
The mean (IQR) 1-year PM2.5, sulfate and ammonium levels were 10.3 (2.15) μg/m3, 3.39 (0.81) μg/m3 and 1.05 (0.29) μg/m3. Supplementary Table 1 presents the Pearson correlation coefficients and the proportion of total PM2.5 mass of GEOS-chem transport model-derived PM2.5 component species across all study visits. Sulfate makes up the greatest proportion of PM2.5 mass (33.2%), and ammonium makes up 10.2% of PM2.5 mass. The correlation coefficients for sulfate and ammonium with total PM2.5 mass are 0.30 (p < 0.0001) and 0.51 (p < 0.0001), respectively.
Elastic net-selected miRNA processing gene SNPs
Supplementary Table 2 lists the 14 miRNA processing SNPs that were included in the elastic net selection model. Of these 14 SNPs, four (rs4961280 [AGO2], rs6877842 [DROSHA], rs910924 [GEMIN4] and rs784567 [TARBP2]) were selected by the elastic net with DNA methylation as the outcome.
Of the four elastic net-selected SNPs, only rs4961280 (AGO2) and rs784567 (TARBP2) were significantly associated with DNAm-age in fully adjusted linear mixed-effects models that included PM2.5 levels as a covariate (Table 2). For rs4961280 (AGO2), individuals who had at least one copy of the minor SNP allele on average had a 1.13-year lower DNAm-age than individuals with the homozygous major variant (allele) genotype (p < 0.05). When we compared all three genotypes, on average, individuals who were homozygous for the minor rs4961280 (AGO2) variant (AA) had the lowest DNAm-age. Homozygous major carriers (CC) had the highest DNAm-age and heterozygous individuals (CA) had an intermediate DNAm-age (Supplementary Figure 1). The trend for this relationship was statistically significant (p = 0.04). For rs784567 (TARBP2), individuals who had at least one copy of the minor SNP allele on average had a 1.35-year lower DNAm-age than individuals with the homozygous major allele genotype (p = 0.04, Table 2). When we looked across all three rs784567 (TARBP2) genotypes, a trend similar to rs4961280 (AGO2) was observed, but the trend did not reach statistical significance (p = 0.08, Supplementary Figure 1). These relationships persisted in sensitivity analyses where the ammonium component was subtracted from total PM2.5 mass (Supplementary Table 3).
Table 2. . Mean 1-year fine particle (PM2.5) concentrations and miRNA processing gene SNPs as joint predictors of DNA methylation age (n = 808).
| Predictor | Difference in DNAm-age for IQR (95% CI) | p-value |
|---|---|---|
| PM2.5 | 0.76 (0.24, 1.24) | 0.003 |
| Elastic net-selected miRNA SNPs† | ||
| rs4961280 (AGO2) | -1.13 (-2.26, -0.002) | 0.05 |
| rs6877842 (DROSHA) | -0.78 (-1.92, 0.37) | 0.18 |
| rs910924 (GEMIN4) | -0.41 (-1.47, 0.65) | 0.45 |
| rs784567 (TARBP2) | -1.35 (-2.61, -0.09) | 0.04 |
Note. Model adjusted for chronological age, blood cell type, temperature, pack years, smoking status, season, BMI, alcohol consumption, education, lifetime cancer diagnosis, hypertension, diabetes and ischemic heart disease.
†Values for the miRNA processing SNPs are in reference to participants whose genotypes are homozygous for the major variant. Bold text specifies statistically significant p-values (< 0.05).
DNAm: DNA methylation; IQR: Interquartile range.
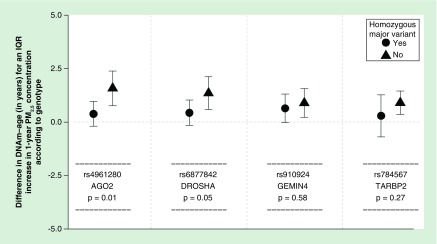
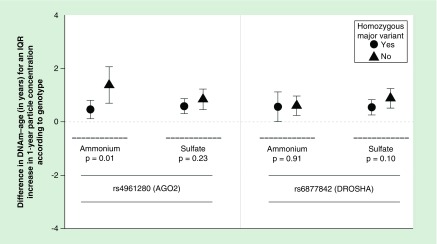
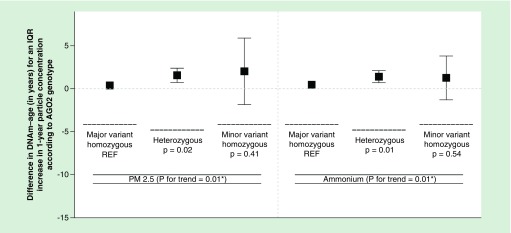
Out of all four SNPs, only the rs4961280 (AGO2) SNP significantly modified the association of PM2.5 with DNAm-age (p = 0.01) – although the rs6877842 (DROSHA) SNP neared statistical significance (p = 0.052, Figure 1). Specifically, the association of PM2.5 with DNAm-age was greater in individuals who were not homozygous for the major AGO2 variant (allele) when compared with individuals who were homozygous for the major allele. This trend was also observed for the ammonium PM2.5 component species. No significant effect modification by the AGO2 SNP was observed for sulfate levels and the rs6877842 (DROSHA) SNP did not significantly modify the relationships of ammonium or sulfate with DNAm-age (Figure 2). We observed a significant (p = 0.01) increasing trend for the association of PM2.5 with DNAm-age when comparing the three rs4961280 (AGO2) SNP genotypes (Figure 3). The strongest (magnitude) association was observed in individuals who were homozygous for the minor allele and the smallest association was observed in individuals with the homozygous major allele genotype. An association of intermediate magnitude was observed in individuals who were heterozygous for the genotype. A similarly significant (p = 0.02) trend across genotypes was observed for the relationship of ammonium with DNAm-age (Figure 3). In a sensitivity analysis where the ammonium component was subtracted from total PM2.5 mass, we still observed a significant – although slightly attenuated – trend in the PM2.5 (less ammonium) and DNAm-age relationship across AGO2 genotypes (Supplementary Figure 2).
Figure 1. . Difference in DNA methylation age for one interquartile range increase in 1-year particle exposure levels comparing participants with and without a homozygous major variant genotype for AGO2, DROSHA, GEMIN4 and TARBP2 in fully adjusted linear mixed-effects models.
Figure 2. . Difference in DNA methylation age for one interquartile range increase in 1-year particle exposure (ammonium and sulfate) levels comparing participants with and without a homozygous major variant genotype for AGO2 and DROSHA in fully adjusted linear mixed-effects models.
Figure 3. . Difference in DNA methylation age for one interquartile range increase in 1-year particle exposure (PM2.5 and ammonium) levels comparing participants of homozygous major variant (n = 526), heterozygous (n = 257) and homozygous minor variant genotypes (n = 25) for AGO2 in fully adjusted linear mixed-effects models.
*p-value for the test of linear trend across genotypes was based on a linear mixed-effects regression model where the three AGO2 genotypes were fit as a continuous measure.
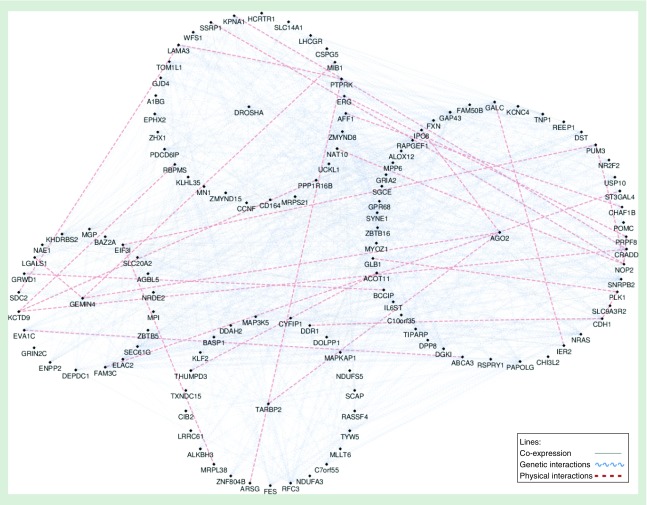
Gene network analysis
Figure 4 depicts the results of a network analysis examining relationships of AGO2, DROSHA, GEMIN4 and TARBP2 with the 353 genes that contribute CpGs to the DNAm-age measure. IPO8 was the sole DNAm-age CpG contributing gene that had a physical interaction with AGO2. Many genes were found to have genetic interactions or be co-expressed with AGO2. PAPOLG and TIPARP were the only two genes that had both genetic interactions and were co-expressed with AGO2. PAPOLG and TIPARP were also co-expressed or had a genetic interaction with DROSHA (Supplementary Table 4).
Figure 4. . Curated network map depicting relationships of AGO2, DROSHA, GEMIN4 and TARBP2 with genes that contribute component CpG methylation to DNA methylation age.
Each of the elastic net-selected genes is surrounded by a circle of related genes that contribute CpG methylation to the DNAm-age metric. Solid lines that connect genes represent co-expression. Dashed lines that connect genes represent physical interactions. Squiggly lines that connect genes represent genetic interactions.
Discussion
The present study utilized a DNAm-age elastic net selection model to identify four SNPs in miRNA processing genes (rs4961280 [AGO2], rs6877842 [DROSHA], rs910924 [GEMIN4] and rs784567 [TARBP2]) of which two (rs4961280 [AGO2] and rs784567 [TARBP2]) were directly associated with DNAm-age in a population of community-dwelling elderly men. Additionally, the study demonstrated a significant modifier effect of the rs4961280 (AGO2) SNP on the associations of 1-year PM2.5 and ammonium (one PM2.5 component species) levels with DNAm-age. More specifically, our data suggest that the association of PM2.5 with DNAm-age is attenuated in individuals carrying at least one copy of the rs4961280 (AGO2) major variant allele. Our results were consistent (though slightly attenuated) in sensitivity analyses where we subtracted ammonium levels from total PM2.5 mass. This suggests that the impact of the rs4961280 (AGO2) SNP on the relationship between PM2.5 and DNAm-age is largely – but not exclusively – due to ammonium levels. Moreover, a gene network analysis revealed physical interactions, genetic interactions and co-expression relationships of AGO2, DROSHA, GEMIN4 and TARBP2 with genes that contribute CpGs to the DNAm-age metric. To our knowledge, this is the first study to examine relationships of miRNA processing physiology with epigenetic age both independently and in the context of long-term PM2.5 exposure.
DROSHA is a gene located on human chromosome 5 and it encodes an RNA-specific endoribonuclease that is involved in the initial step of nuclear miRNA processing [36]. GEMIN4 and TARBP2 are located on chromosomes 17 and 12, respectively, and they both encode enzymes that are involved in the cytoplasmic processing of miRNAs. After a literature review examining the relationships of the SNPs in these three genes with air pollution, we found only one previous study – also in the Normative Aging Study cohort – demonstrating that in comparison to other participants, individuals heterozygous for the rs910924 (GEMIN4) SNP genotype were more likely to have lower global cognition measurements given the same level of black carbon exposure [24]. Nonetheless, we did find a number of studies implicating DROSHA, GEMIN4 and TARBP2 in numerous aging-related diseases including prostate cancer and colorectal cancer [37,38]. Since cancers are often related to changes in DNA methylation, the connections of these genes with cancer may be the reason why their SNPs were selected by the elastic net [39].
In contrast to the other three SNPs, we did observe significant effect modification of the PM2.5–DNAm-age relationship by the rs4961280 (AGO2) SNP. Furthermore, we also observed a significant direct relationship of the rs4961280 (AGO2) SNP with DNAm-age. AGO2 (Argonaute Protein 2) is a gene located on human chromosome 2. Argonaute (AGO) proteins, including AGO2, form the core of the RNA-induced silencing complex (RISC) which is involved in gene silencing via RNAi. AGO proteins are well conserved across species and structurally include an amino-terminal, PAS, Piwi, and MID domains. Humans have eight AGO proteins; however, only AGO 1–4 are capable of loading miRNA in RISC. Moreover, only AGO2 appears to have the ability to cleave mRNA targets and achieve transcript instability/silencing [40]. In addition to their role in RISC, it has also been demonstrated that AGO proteins play a role in stabilizing and maintaining proper levels of mature miRNA strands [41].
Our data suggested that individuals with at least one copy of the rs4961280 (AGO2) major variant had an attenuated association of PM2.5/ammonium with DNAm-age when compared with individuals who were homozygous for the minor variant. However, individuals with at least one copy of the minor variant on average had lower DNAm-ages when compared with individuals that were homozygous for the major variant. Very few studies have explicitly examined relationships of the rs4961280 (AGO2) SNP and none of them were in the contexts of PM2.5 or aging. However, we did find one study that demonstrated that the minor variant was associated with a reduced risk of benign prostatic hypertrophy (BPH) in a Serbian population [37]. Since BPH is most common in aging men, this study is in alignment with our finding that the minor allele is associated with qualities of being ‘younger’ (i.e., a lower DNAm-age or a lower risk of BPH) [42]. Further work will need to be done to understand why it is the major allele that attenuates the positive association of PM2.5 with DNAm-age, but the minor allele that is directly associated with a lower biological age. Moreover, it will be helpful for the field to understand why ammonium relationships with DNAm-age were impacted by the rs4961280 (AGO2) SNP and sulfate relationships were not.
Of the existing studies that examine relationships of AGO2 – not simply the SNP, but the gene – with aging in humans, the majority are cell culture based and describe AGO2 as a factor involved in molecular processes related to biological aging such as cellular senescence, stem cell renewal and endothelial function [43–45]. We also found a few animal studies that showed relationships of AGO2 with chronological aging. A study examining relationships of miRNAs with aging in Drosophila revealed that with age, there was a global increase in miRNAs loaded in AGO2 but not AGO1. Furthermore, mutations in AGO2 resulted in shorter life span and neurodegeneration. Together, these data suggest that AGO2 impacts aging-associated processes [46]. Another study looking to elucidate how intermittent fasting increases longevity in Caenorhabditis elegans demonstrated that fasting upregulates the expression of miRNA-induced silencing complex (RISC) components including AGOs. In this study, fasting upregulated AGO2 by twofold [47].
Our network analyses of AGO2, DROSHA, GEMIN4, TARBP2 and genes that contribute CpGs to the DNAm-age metric demonstrated one physical interaction between AGO2 and a gene called IPO8. IPO8 (Importin 8) is a gene on chromosome 12 that encodes a protein involved in mediating the nuclear import of other proteins with nuclear localization signals. IPO8 has also been shown to mediate the cytoplasm to nucleus transport of mature miRNAs. Moreover, this IPO8-mediated transport of miRNAs is dependent on the physical association of IPO8 with the AGO2 complex [48]. The literature primarily describes IPO8 as an optimum reference gene for microarray and RT-PCR studies in multiple tissue types including the lung [49]. Two genes (PAPOLG and TIPARP) had both genetic interactions and co-expression relationships with AGO2. These two genes also had genetic interactions with TARBP2 and DROSHA. PAPOLG (Poly [A] polymerase γ) is a gene on chromosome 2 that encodes an exclusively nuclear-localized poly (A) polymerase responsible for catalyzing template-independent extension of the 3′ end of a strand of DNA/RNA [50]. To our knowledge, no explicit studies have examined the relationships of PAPOLG and biological aging but PAPOLG has been implicated in relationships involving aging-related health outcomes. For instance, a 12-week trial examined if a dietary intervention of 400 g/week of high-glucoraphanin broccoli altered plasma metabolites linked to cancer risk when compared with diets of 400 g/week of standard broccoli or 400 g/week of peas. No other modifications were made to the participants’ diets. The study revealed that the levels of plasma metabolites (including flavin adenine dinucleotide [FAD]) of individuals receiving high-glucoraphanin broccoli were differentiated by PAPOLG genotypes. This suggests that PAPOLG may interact with diet to impact the levels of metabolites including those that have been implicated in cancer risk [51]. TIPARP (tetrachlorodibenzo-p-dioxin-inducible poly [ADP-ribose] polymerase) is a gene on chromosome 3 that encodes a member of the poly (ADP-ribose) polymerase super family [52]. In a study exposing human aortic endothelial cells to 10 μg/ml of fine and ultrafine ambient particulate matter from California, mRNA levels of enzymes including TIPARP increased [53]. Another study exposed human adenocarcinomic human alveolar basal epithelial (A549) cells to 10 μg/ml of winter and summer PM2.5 from Milan and found that PM2.5 from both seasons modulated TIPARP gene expression [54].
Strengths of our study include the combination of a novel biomarker, rigorous statistical methods and access to a large cohort with extensive and repeated information regarding PM2.5 exposure levels, DNA methylation data and potential confounders from multiple study visits. This is the first study to use miRNA processing gene variants to study the relationship of ambient fine particles with DNAm-age. However, our study does have some notable limitations. First, we use a validated chemical transport model to generate address-level 1-year PM2.5, sulfate, and ammonium exposure estimates. Given that most NAS participants are retired, we believe that particle exposure levels at their homes approximately capture their personal exposures. Still, there is some risk of exposure misclassification. Nonetheless, such nondifferential misclassification is likely to underestimate any observed associations rather than bias them away from the null [55]. Second, we utilize a panel of a miRNA processing gene SNPs that is somewhat limited because it does not provide genome-wide resolution of all genes involved in miRNA processing. Nevertheless, this panel has been successfully utilized in other environmental health studies and we use a rigorous elastic net approach to identify our variants of interest [27]. Although we did not test for effect modification with all the SNPs in our panel, our targeted approach identified significant interactions that persisted even in sensitivity analyses. These findings will be informative to more comprehensive, future research. Last, our study examines the role of the miRNA processing pathway by using genetic variants of miRNA processing genes and our findings are based on a cohort of elderly Caucasian males who reside in a lightly polluted environment. Future studies involving other demographic groups, in different environments and using miRNA expression levels will be necessary to broadly confirm and add to these important but early findings.
Conclusion
In conclusion, genotypes of the rs4961280 (AGO2) miRNA processing SNP were directly associated with DNAm-age and modified the associations of 1-year PM2.5 and ammonium levels with DNAm-age in this population of community-dwelling Caucasian elderly men. Although our findings need to be confirmed in other individuals of this same demographic group and different populations, they begin to address the important research gap concerning the biological relevance of DNAm-age and the physiology of the PM2.5–DNAm-age relationship. Future studies will be necessary to elucidate more nuanced relationships of miRNA physiology with epigenetic aging.
Summary points.
Since miRNAs are heavily involved in gene regulation, we investigated the modifying role of genetic variation in miRNA processing genes on the PM2.5–DNA methylation (DNAm)-age relationship.
After identifying SNPs important to DNAm-age with an elastic net, we tested for PM2.5–SNP interactions associated with DNAm-age.
Four SNPs (rs4961280 [AGO2], rs6877842 [DROSHA], rs910924 [GEMIN4] and rs784567 [TARBP2]) were selected by the elastic net.
Only the rs4961280 [AGO2] SNP modified the PM2.5–DNAm-age relationship.
In fully adjusted linear mixed-effects models, the association of PM2.5 with DNAm-age was significantly (Pinteraction = 0.01) weaker in homozygous carriers of the major AGO2 allele (β = 0.38; 95% CI: -0.20, 0.96) when compared with all other participants (β = 1.58; 95% CI: 0.76, 2.39).
A gene network analysis revealed known physical, genetic and co-expression relationships of AGO2 with genes that contribute methylation values to the DNAm-age measure including IPO8, PAPOLG and TIPARP.
We observed no effect modification by the other SNPs.
Our results suggest that miRNA processing impacts DNAm-age relationships particularly in the context of long-term PM2.5 exposure.
Supplementary Material
Footnotes
Data availability
Data are from the Normative Aging Study, from which restricted data are available for researchers who meet the criteria. Methylation data are deposited at NCBI dbGaP (study accession number: phs000853.v1.p1).
Financial & competing interests disclosure
This work was supported by grants from the National Institute of Environmental Health Sciences (NIEHS) (R01ES021733 and R01ES025225). Other support comes from NIEHS grants (ES015172, ES014663, ES020010, P30ES009089 and P30ES000002); Environmental Protection Agency grants (RD832416 and RD83587201); and a National Heart, Lung, and Blood Institute grant (2T32HL007118-41). Additional support was provided by the US Department of Agriculture, Agricultural Research Service (contract 53-K06–510). The US Department of Veterans Affairs Normative Aging Study is supported by the Cooperative Studies Program/ERIC, US Department of Veterans Affairs, and is a research component of the Massachusetts Veterans Epidemiology Research and Information Center. The views expressed in this paper are those of the authors and do not necessarily represent the views of the US Department of Veterans Affairs. JC Nwanaji-Enwerem is also supported by an NIH/NIA Ruth L Kirschstein National Research Service Award (1 F31AG056124–01A1). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethics approval
Boston VA Medical Center, Harvard TH Chan School of Public Health (protocol 14027–102).
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.WhO. Ageing and health. 2015. www.who.int/ageing/events/world-report-2015-launch/en/
- 2.Association As. 2017 Alzheimer's disease facts and figures. Alzheimer's & Dementia. 2017:13325–13373. www.alz.org/documents_custom/2017-facts-and-figures.pdf In. [Google Scholar]
- 3.WHO. WHO; Geneva: 2014. Burden of disease from household air pollution for 2012.www.who.int/phe/health_topics/outdoorair/databases/FINAL_HAP_AAP_BoD_24March2014.pdf [Google Scholar]
- 4.Zhong J, Cayir A, Trevisi L, et al. Traffic-related air pollution, blood pressure, and adaptive response of mitochondrial abundance. Circulation. 2016;133(4):378–387. doi: 10.1161/CIRCULATIONAHA.115.018802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schikowski T, Vossoughi M, Vierkotter A, et al. Association of air pollution with cognitive functions and its modification by APOE gene variants in elderly women. Environ. Res. 2015;142:10–16. doi: 10.1016/j.envres.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Raaschou-Nielsen O, Beelen R, Wang M, et al. Particulate matter air pollution components and risk for lung cancer. Environ. Int. 2016;87:66–73. doi: 10.1016/j.envint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Nwanaji-Enwerem JC, Colicino E, Trevisi L, et al. Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ. Epigenet. 2016;2(2):dvw006. doi: 10.1093/eep/dvw006. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first published work reporting associations of PM2.5 with DNA methylation (DNAm) age.
- 8.Nwanaji-Enwerem JC, Dai L, Colicino E, et al. Associations between long-term exposure to PM2.5 component species and blood DNA methylation age in the elderly: the VA Normative Aging Study. Environ. Int. 2017;102:57–65. doi: 10.1016/j.envint.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8(9):1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A key study that presents data suggesting that DNAm-age is an important measure of mortality risk.
- 10.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging (Albany NY). 2015;7(12):1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first report of the DNAm-age measure that we are using and it discusses many of the basic properties of the measure.
- 12.Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. doi: 10.1186/s13072-017-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodosthenous RS, Coull BA, Lu Q, et al. Ambient particulate matter and microRNAs in extracellular vesicles: a pilot study of older individuals. Part Fibre Toxicol. 2016;13:13. doi: 10.1186/s12989-016-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Vijayan M, Bhatti JS, Reddy PH. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog. Mol. Biol. Transl. Sci. 2017;146:47–94. doi: 10.1016/bs.pmbts.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Parodi F, Carosio R, Ragusa M, et al. Epigenetic dysregulation in neuroblastoma: a tale of miRNAs and DNA methylation. Biochim. Biophys. Acta. 2016;1859(12):1502–1514. doi: 10.1016/j.bbagrm.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Morita S, Horii T, Kimura M, et al. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89(6):687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Hodjat M, Rahmani S, Khan F, et al. Environmental toxicants, incidence of degenerative diseases, and therapies from the epigenetic point of view. Arch. Toxicol. 2017;91(7):2577–2597. doi: 10.1007/s00204-017-1979-9. [DOI] [PubMed] [Google Scholar]
- 18.Bell B, Rose CL, Damon A. The veterans administration longitudinal study of healthy aging. Gerontologist. 1966;6(4):179–184. doi: 10.1093/geront/6.4.179. [DOI] [PubMed] [Google Scholar]; • Important for understanding the Normative Aging Study cohort.
- 19.DNA methylation age and theepigenetic clock. http://labs.genetics.ucla.edu/horvath/dnamage/
- 20.GEOS-Chem Model. www.geos-chem.org
- 21.Di Q, Koutrakis P, Schwartz J. A hybrid prediction model for PM2.5 mass and components using a chemical transport model and land use regression. Atmospheric Environment. 2016;131:390–399. [Google Scholar]
- 22.Yang H, Dinney CP, Ye Y, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68(7):2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 23.Horikawa Y, Wood CG, Yang H, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 2008;14(23):7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colicino E, Giuliano G, Power MC, et al. Long-term exposure to black carbon, cognition and single nucleotide polymorphisms in microRNA processing genes in older men. Environ. Int. 2016;88:86–93. doi: 10.1016/j.envint.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilker EH, Baccarelli A, Suh H, et al. Black carbon exposures, blood pressure, and interactions with single nucleotide polymorphisms in microRNA processing genes. Environ. Health Perspect. 2010;118(7):943–948. doi: 10.1289/ehp.0901440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.PhAT. Pharmacogenetics of Asthma Treatment. www.pharmgat.org/Tools/pbtoldplotform
- 27.Lenters V, Portengen L, Rignell-Hydbom A, et al. Prenatal phthalate, perfluoroalkyl acid, and organochlorine exposures and term birth weight in three birth cohorts: multi-pollutant models based on elastic net regression. Environ. Health Perspect. 2016;124(3):365–372. doi: 10.1289/ehp.1408933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.glmnet: Lasso and Elastic-Net Regularized Generalized Linear Models. https://cran.r-project.org/web/packages/glmnet/index.html
- 29.Nwanaji-Enwerem JC, Bind MA, Dai L, et al. Modifying role of endothelial function gene variants on the association of long-term PM2.5 exposure with blood DNA methylation age: the VA Normative Aging Study. Toxicol. Sci. 2017;158(1):116–126. doi: 10.1093/toxsci/kfx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 31.Zou H, Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. 2005;67(2):301–320. [Google Scholar]
- 32.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panni T, Mehta AJ, Schwartz JD, et al. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the Normative Aging Study. Environ. Health Perspect. 2016;124(7):983–990. doi: 10.1289/ehp.1509966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai L, Mehta A, Mordukhovich I, et al. Differential DNA methylation and PM2.5 species in a 450 K epigenome-wide association study. Epigenetics. 2016;12(2):139–148. doi: 10.1080/15592294.2016.1271853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GeneMANIA. https://genemania.org
- 36.Daugaard I, Hansen TB. Biogenesis and function of Ago-associated RNAs. Trends Genet. 2017;33(3):208–219. doi: 10.1016/j.tig.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Nikolic Z, Savic Pavicevic D, Vucic N, et al. Genetic variants in RNA-induced silencing complex genes and prostate cancer. World J. Urol. 2017;35(4):613–624. doi: 10.1007/s00345-016-1917-0. [DOI] [PubMed] [Google Scholar]
- 38.Mullany LE, Herrick JS, Wolff RK, Buas MF, Slattery ML. Impact of polymorphisms in microRNA biogenesis genes on colon cancer risk and microRNA expression levels: a population-based, case–control study. BMC Med. Genomics. 2016;9(1):21. doi: 10.1186/s12920-016-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.An J, Rao A, Ko M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med. 2017;49(4):e323. doi: 10.1038/emm.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meister G, Landthaler M, Patkaniowska A, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Azhar G, Wei JY. The expression of microRNA and microRNA clusters in the aging heart. PLoS ONE. 2012;7(4):e34688. doi: 10.1371/journal.pone.0034688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unnikrishnan R, Almassi N, Fareed K. Benign prostatic hyperplasia: evaluation and medical management in primary care. Cleve Clin. J. Med. 2017;84(1):53–64. doi: 10.3949/ccjm.84a.16008. [DOI] [PubMed] [Google Scholar]
- 43.Kim BS, Im YB, Jung SJ, Park CH, Kang SK. Argonaute2 regulation for K+ channel-mediated human adipose tissue-derived stromal cells self-renewal and survival in nucleus. Stem Cells Dev. 2012;21(10):1736–1748. doi: 10.1089/scd.2011.0388. [DOI] [PubMed] [Google Scholar]
- 44.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 2012;14(3):266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez P, Wagner KD, Hofman P, Van Obberghen E. RNA activation of the vascular endothelial growth factor gene (VEGF) promoter by double-stranded RNA and hypoxia: role of noncoding VEGF promoter transcripts. Mol. Cell Biol. 2016;36(10):1480–1493. doi: 10.1128/MCB.01096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abe M, Naqvi A, Hendriks GJ, et al. Impact of age-associated increase in 2′-O-methylation of miRNAs on aging and neurodegeneration in Drosophila . Genes Dev. 2014;28(1):44–57. doi: 10.1101/gad.226654.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kogure A, Uno M, Ikeda T, Nishida E. The microRNA machinery regulates fasting-induced changes in gene expression and longevity in Caenorhabditis elegans . J. Biol. Chem. 2017;292(27):11300–11309. doi: 10.1074/jbc.M116.765065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinmann L, Hock J, Ivacevic T, et al. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136(3):496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 49.Nguewa PA, Agorreta J, Blanco D, et al. Identification of importin 8 (IPO8) as the most accurate reference gene for the clinicopathological analysis of lung specimens. BMC Mol. Biol. 2008;9:103. doi: 10.1186/1471-2199-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kyriakopoulou CB, Nordvarg H, Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J. Biol. Chem. 2001;276(36):33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- 51.Armah CN, Traka MH, Dainty JR, et al. A diet rich in high-glucoraphanin broccoli interacts with genotype to reduce discordance in plasma metabolite profiles by modulating mitochondrial function. Am. J. Clin. Nutr. 2013;98(3):712–722. doi: 10.3945/ajcn.113.065235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh M, Katoh M. Identification and characterization of human TIPARP gene within the CCNL amplicon at human chromosome 3q25.31. Int. J. Oncol. 2003;23(2):541–547. [PubMed] [Google Scholar]
- 53.Aung HH, Lame MW, Gohil K, et al. Comparative gene responses to collected ambient particles in vitro: endothelial responses. Physiol. Genomics. 2011;43(15):917–929. doi: 10.1152/physiolgenomics.00051.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gualtieri M, Longhin E, Mattioli M, et al. Gene expression profiling of A549 cells exposed to Milan PM2.5 . Toxicol. Lett. 2012;209(2):136–145. doi: 10.1016/j.toxlet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 55.Kioumourtzoglou MA, Spiegelman D, Szpiro AA, et al. Exposure measurement error in PM2.5 health effects studies: a pooled analysis of eight personal exposure validation studies. Environ. Health. 2014;13(1):2. doi: 10.1186/1476-069X-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.