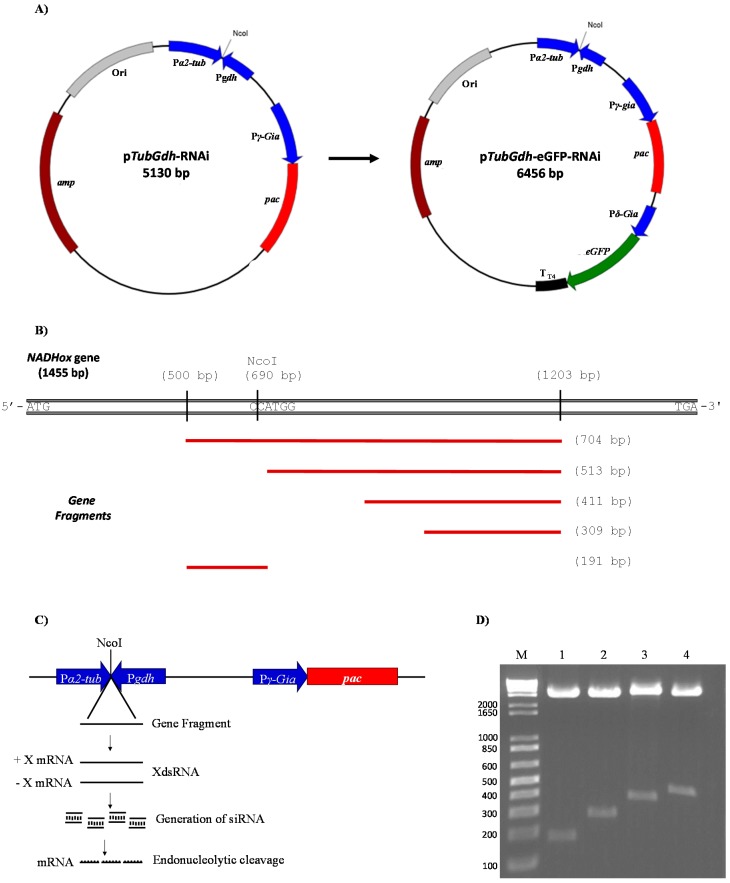
Figure 1.
Vectors and strategy to generate double-stranded RNAs (dsRNAs) from the NADHox gene. (A) Map of the pTubGdh-RNAi and pTubGdh_eGFP-RNAi vectors, showing the position of the α2-Tub::gdh cassette with the NcoI cloning site. The pac gene, conferring resistance to puromycin, was used as selective marker for transformation. (B) Position of the fragments used for gene silencing of the NADHox gene. (C) Close-up of the α2-Tub::gdh cassette, showing the expected outcome after insertion of the NADHox gene fragments at the NcoI restriction site, with the generation of dsRNA from the corresponding fragment and subsequent silencing of NADHox expression through the RNAi pathway. (D) The insertion of the NADHox gene fragments into the vectors was confirmed by digestion with NcoI and electrophoresis; lanes 1–4 show the released fragments of approximately 200, 300, 400 and 500 base pair (bp) from vector pTubGdh_eGFP-RNAi, respectively, and lane M is the molecular weight marker (1 Kb Ladder, Invitrogen).