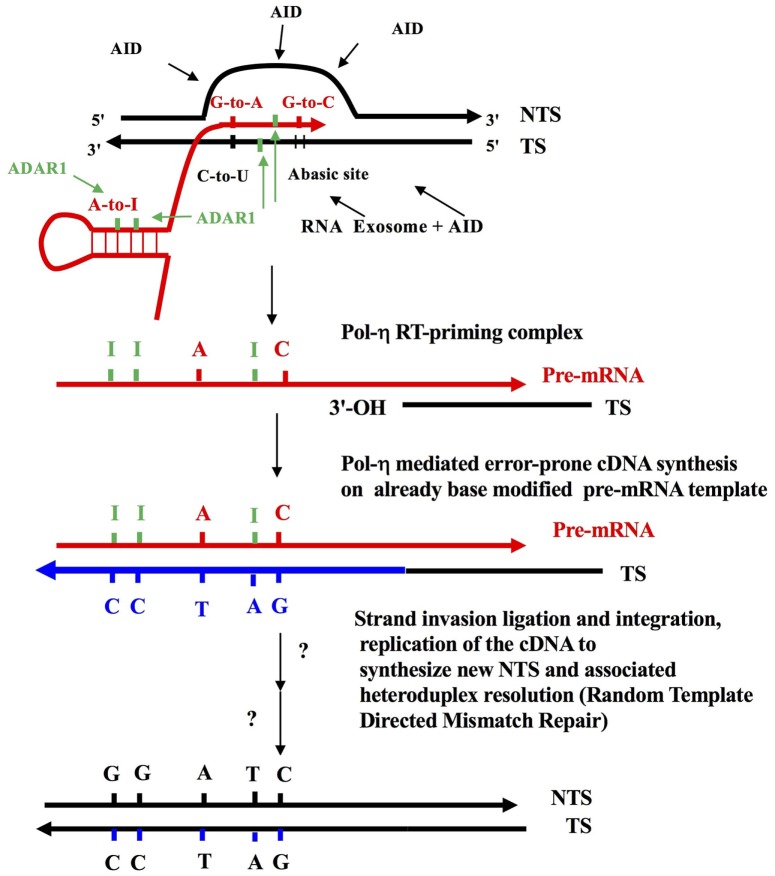
Figure 1.
The reverse transcriptase mechanism of somatic hypermutation (SHM). Some elements of this figure have appeared before, and this figure in toto is a modified combination of parts from Figure 1 in Lindley and Steele (10), as well as from figures in Steele (9, 11) and Steele and Lindley (14). This is also an adaptation of the target site reverse transcription process of Luan et al. (66). Shown is an RNA Polymerase II-generated Transcription Bubble with C-site and A-site substrate deamination events by AID and ADAR proteins, which generates the strand-biased mutation signatures—A-to-G, G-to-A, G-to-T, and G-to-C (9, 11, 14). DNA strands shown by black lines; pre-mRNA as red lines; cDNA strands as thick blue lines due to DNA polymerase η (59). Green bars are Inosines. Shown also is the action of the RNA exosome (64) allowing access of AID deaminase to cytosines on the transcribed strand (TS). The ssDNA regions on the displaced non-transcribed strand (NTS) are established targets of AID action (53–56). Note that DNA mutations are first introduced as AID-mediated C-to-U, followed by excision of uracils by DNA glycosylase (UNG), which creates Abasic sites in the TS (these can mature into single strand nicks with 3′-OH ends via the action of AP endonuclease). These template Uracil and Abasic sites can be copied into pre-mRNA by RNA Pol II generating G-to-A and G-to-C modifications as shown (67). Following target site reverse transcription (66), this results in G-to-A and G-to-C mutations in the NTS, in a strand biased manner (9–11, 14). Separately at WA targets in nascent dsRNA substrates, adenosine-to-inosine (A-to-I) RNA editing events, mediated by ADAR1 deaminase, are copied back into DNA by reverse transcription via Pol-η (59). In theory, ADARs can also deaminate the RNA and DNA moieties in the RNA: DNA hybrid (14, 15). The strand invasion and integration of newly synthesized cDNA TS, as well as random-template mismatch repair (68) are hypothesized additional steps (not shown here). In short, RNA Pol II introduces modifications in the Ig pre-mRNA as it copies TS DNA with AID lesions and this is coupled to A-to-I in dsRNA stem-loops near the transcription bubble (62) as well as in RNA:DNA hybrids within the bubble (14, 15). Next, a RT-priming substrate is formed when the nicked TS strand with an exposed 3′-OH end anneals with the base modified pre-mRNA copying template allowing cDNA synthesis by Y Family translesion DNA polymerase-η (48), now acting in its reverse transcriptase mode (59). These 3′-OH annealed priming sites could arise due to excisions at previous AID-mediated Abasic sites. Alternatively, they could arise due to an endonuclease excision associated with the MSH2-MSH6 heterodimer engaging a U:G mispaired lesion (61). Shown is an A-to-T transversion generated at the RT step at a template Inosine. ADAR, Adenosine Deaminase that acts on RNA; AP, an Abasic, or apurinic/apyrimidinic, site; APOBEC family, generic abbreviation for the dC-to-dU deaminase family of which AID is a member (e.g., APOBEC1; APOBEC3 A, B, C, D, F, G, H); AID, activation induced cytidine deaminase causing C-to-U lesions at WRCY/RGYW C-site motifs in ssDNA; W, A, or U/T; WA-site, target motif for ADAR deaminase including DNA polymerase-η error prone incorporation in vitro (50, 51); Y, pyrimidines T/U or C.; R, purine A or G.