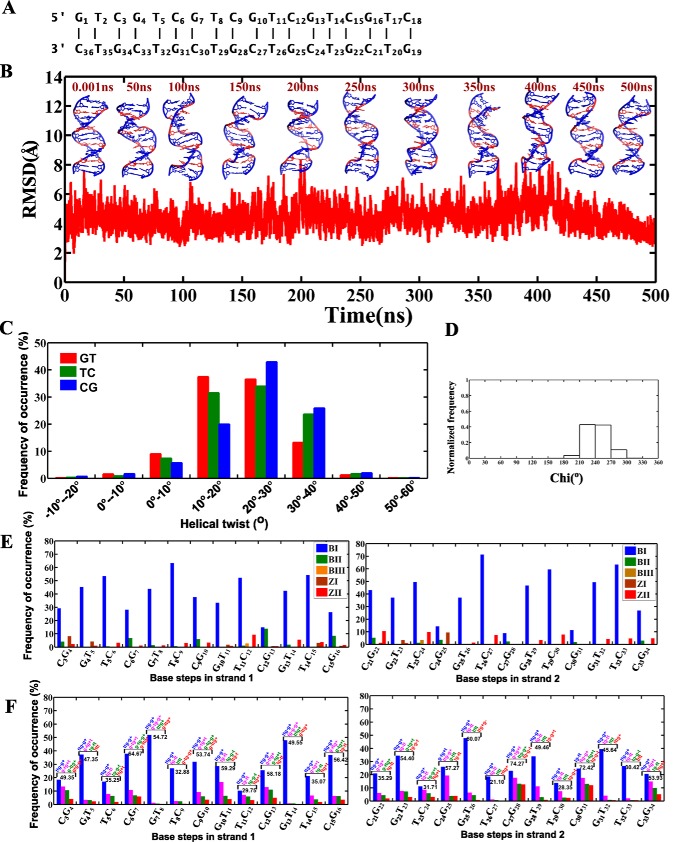
Figure 5.
The d(GTC)6·d(GTC)6 duplex containing the T … T mismatch retains B-form geometry. A, sequence of the 18-mer DNA duplex containing the T … T mismatch used for MD simulation. B, time versus RMSD profile along with a schematic of the d(GTC)6·d(GTC)6 duplex at various time intervals. C, histograms corresponding to helical twists at GT, TC, and CG steps over the last 400 ns. D, glycosyl chi angle values for guanine residues (favoring the anti/high-anti conformation). E, backbone conformational (ϵ,ζ,α,γ) angles for canonical base pairs over the last 400 ns of simulations. F, frequency of occurrence of the top four populations other than BI, BII, BIII, ZI, and ZII are given for each base step. The d(GTC)6·d(GTC)6 sequences were carried out using pmemd.cuda of the AMBER 16 suite.