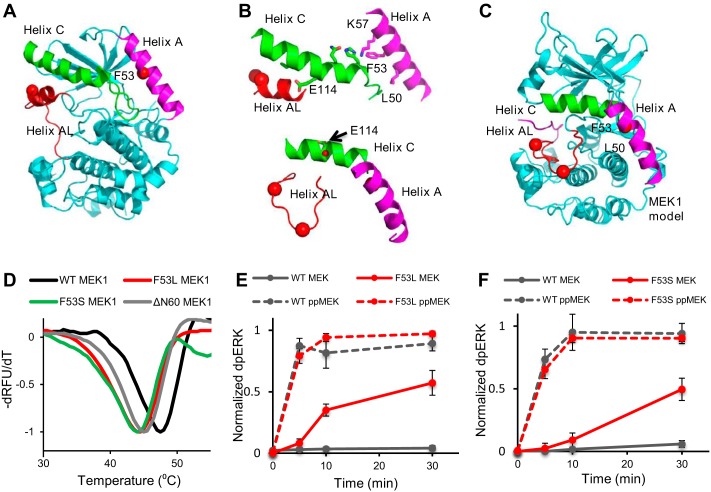
Figure 1.
Activating mutations affect thermal stability of MEK1 and its activity toward ERK2. A, location of position 53 in helix A from the structure of MEK1 in PDB code 5bx0 is indicated by a red dot. Shown are helix A (magenta), helix C (green), and helix AL (red), an inhibitory helix in the activation loop. B, top, close connection between helix A, helix C, and helix AL is shown in inactive MEK1. MEK1 is rotated 180° from A. Side chains involved in ionic interactions along helix C are shown. Phe-53 stabilizes these interactions through Lys-57. Glu-114, a catalytic residue, interacts with helix AL. Bottom, model of the three helices in the active configuration of MEK1 based on the structure of active PKA (PDB code 1ATP). Helix AL is refolded, building the active site. Glu-114 points back, adopting interactions in the active site (red dot, Glu-114). C, model for active MEK1, the whole protein based on PKA (PDB code 1ATP). Helix A adopts different interactions with helix C. D, thermal stability of wild-type MEK1 (black), F53L (red), and F53S (green) and ΔΝ60 (eliminating helix A) (gray) as visualized with SYPRO Orange dye. E and F, MEK1/F53L and MEK1/F53S are intrinsically active and, upon phosphorylation, become as active as phosphorylated wild-type MEK1. The normalized dpERK level is an average of four experimental replicates in E and F. Error bars, S.E.