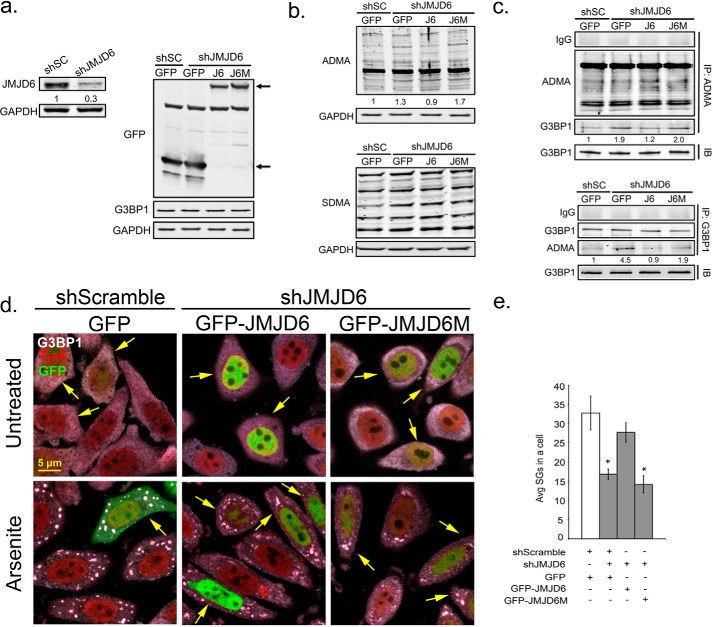
Figure 6.
JMJD6 is a potential demethylase for G3BP1 to promote SG formation. Shown is reconstitution of JMJD6 by wild-type GFP-JMJD6 (J6) or catalytic mutant GFP-JMJD6 (J6M) during knockdown of endogenous JMJD6 in HeLa cells. a, immunoblot analysis for validation of JMJD6 knockdown efficiency (left panel) and GFP, J6, or J6M transgene expression levels (right panel) gene products marked by arrows. b, antibodies specific for asymmetric (ADMA) methyl modifications (top panel), and symmetric methyl modifications (SDMA) (bottom panel) were used in Western blot to examine methylation levels of total endogenous proteins in JMJD6 rescued cells. Relative densitometric analysis of total ADMA signal is shown below immunoblot. c, immunoprecipitation with ADMA antibodies (top panel) or G3BP1 antibodies (bottom panel) to analyze ADMA levels on G3BP1. d, SGs were induced by arsenite and visualized in HeLa cells under the same treatment conditions. SG markers are G3BP1 (gray) and Tia1 (red). Arrows in the panels indicate transfected cells. e, quantification of average SGs/cell in JMJD6 rescued HeLa cells. The white bar indicates shSC control cells, and gray bars indicate JMJD6 knockdown cells. *, p < 0.05 versus shSC control. The results shown are representative of three independent experiments that were conducted in which 100 cells were counted in each for IFA. Original magnification was 63×.