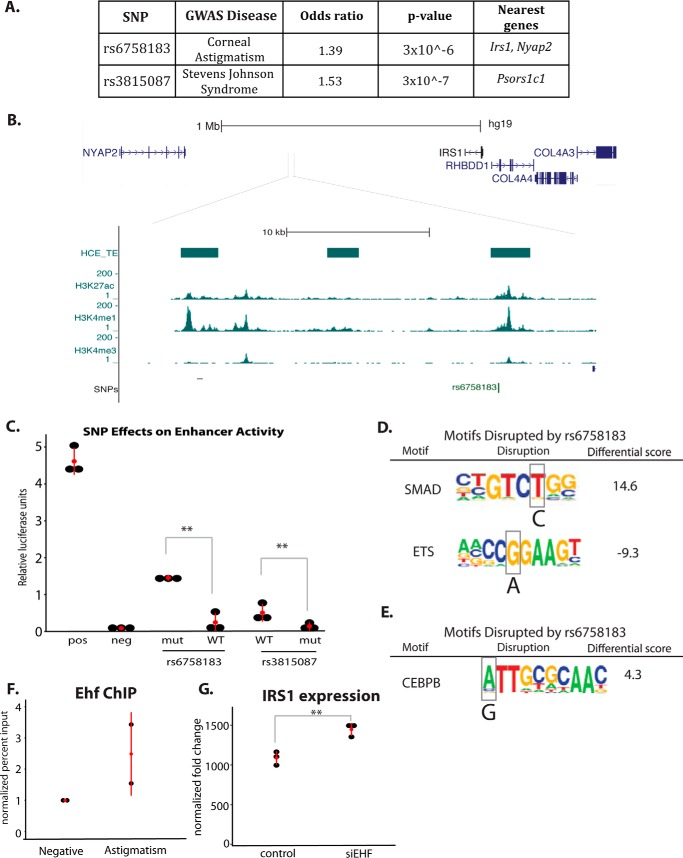
Figure 7.
HCE cell enhancers overlap SNPs linked to corneal diseases. A, table of GWAS SNP information. B, genomic view of rs6758183 with HCE enhancer data. C, luciferase reporter assays for WT and mutant (disease-associated allele) SNPs (n = 6, t test). D, analysis of motif disruption by SNP rs6758183. Scores were calculated by TRAP (24): differential score = mutant score − WT score. E, motif disruption analysis for rs3815087. Scores were calculated by TRAP. F, EHF ChIP in HCE cells at the rs6758183 (astigmatism) region compared with a negative control region. G, IRS1 expression in siRNA control and siEHF-treated HCE cells, detected by microarray as in Ref.18 (n = 3; *, p < 0.05; **, p < 0.01.