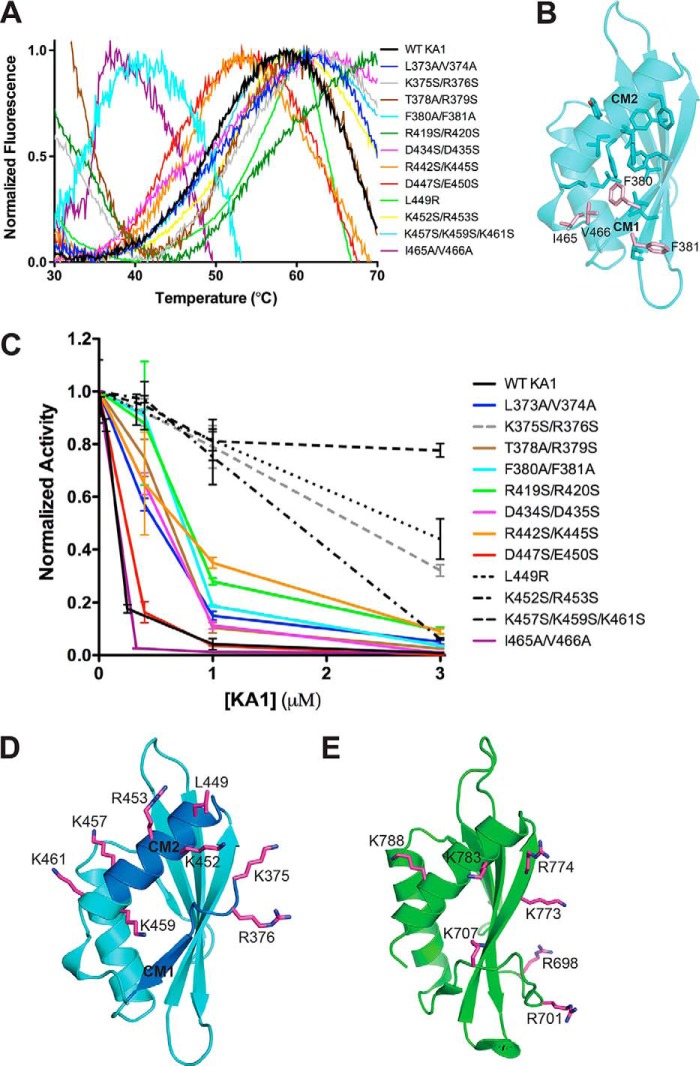
Figure 5.
The autoinhibitory face of Chk1 KA1 domains includes CM1 and CM2. A, melting curves of Chk1 KA1 domain mutants at 25 to 50 μm based on SYPRO Orange fluorescence. All melting temperatures cluster from ∼45 to ∼55 °C except for F380A/F381A and I465A/V466V, which show reduced melting temperatures of ∼35 °C. The K365S/R376S, T378A/R379S, and R419S/R420S mutants likely show high fluorescence readings at 30 °C due to SYPRO Orange binding to exposed hydrophobic protein patches prior to temperature-induced protein unfolding. B, residues contributing to the hydrophobic core of the Chk1 KA1 domain (sticks) include Phe-380, Ile-465, and Val-466 (salmon sticks). C, Chk1 kinase domain (1–277) at 0.05 μm was assayed in the presence of up to 3 μm of various surface-mutated KA1 domain (366–476) constructs compared with WT (black line) with mean ± S.D. of three replicates plotted. Lines connecting the data points are included for clarity. Mutants that most significantly abrogated inhibitory activity (dashed lines) were located on or near CM1 (gray) or CM2 (black). D, residues most critical for KA1-mediated inhibition (magenta sticks) are mapped onto the Chk1 KA1 domain structure (cyan schematic) with the location of CM1 and CM2 indicated (blue). Because these were not part of the crystal construct, Lys-375 and Arg-376 were modeled in COOT (39) as was the side chain of Lys-457, which was disordered in the crystal structure. E, residues (magenta sticks) of the MARK1 KA1 domain (green schematic) previously implicated in autoinhibition (16).