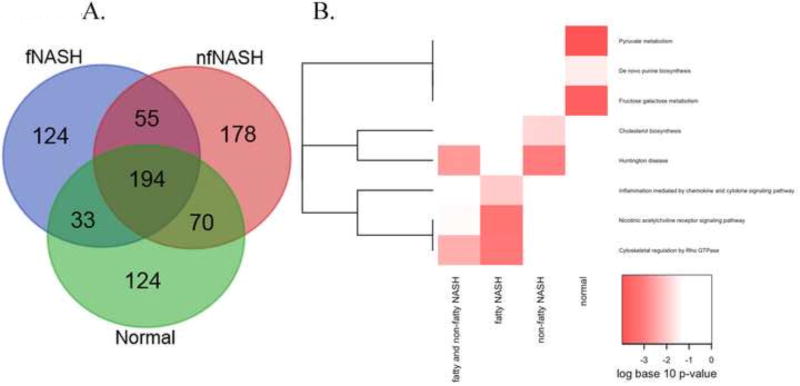
Figure 6. Bioinformatic analysis of carbonylated proteins in end-stage NASH.
A. Proteins identified in Supplemental Table S1 were analyzed using the VENN data analysis software at the Bioinformatics and Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/). B. For enrichment analysis, four groups of proteins were identified. Proteins were functionally annotated and pathways were examined for enrichment using EnrichR. Pathways that were nominally significant (p<0.01) in at least one of the 4 protein lists are included in the graphic. The colors of the heatmap range from white (unadjusted p-value>0.01) to bright red based on the log base 10 transformation of the unadjusted p-value. A p-value of 1 was used when the pathway was not represented by any proteins in the list. PANTHER pathways (rows) are ordered based on hierarchical clustering using the Euclidean distance and a binary indicator of significance (p<0.01 vs. p>=0.01).