Figure 2. Characterization of pH/H2O2-activatable NIR fluorescent nanoprobes.
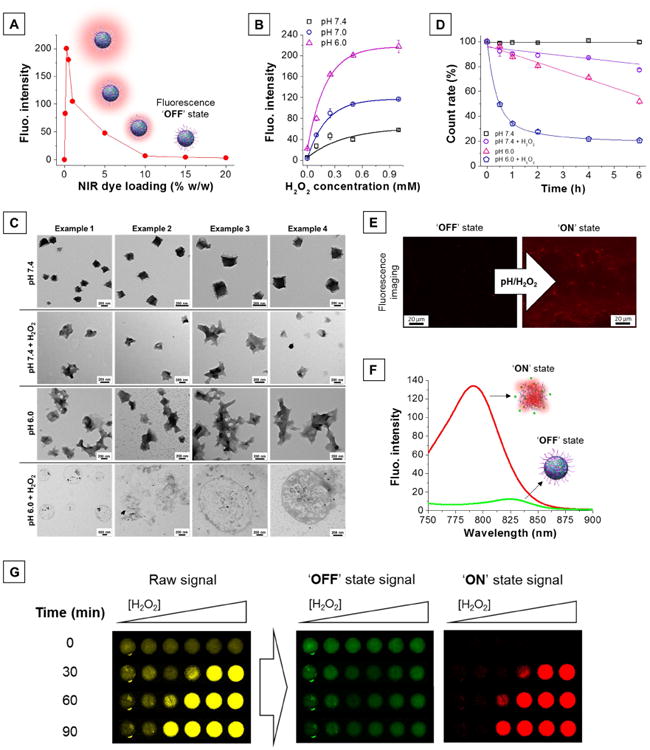
(A) High dye loading in nanoprobes results in quenching of their fluorescence properties, creating an ‘OFF’ state. (B) H2O2-triggered fluorescence activation of dual-responsive nanoprobes is enhanced at low pH. (C) Transmission electron micrographs of the nanoprobes in different environments (pH 7.4, pH 7.4 + H2O2, pH 6.0, pH 6.0 + H2O2). [H2O2] = 0.25 mM, T = 37°C, incubation time: 2 h. (D) Number of nanoprobes over time in different environments (pH 7.4, pH 7.4 + H2O2, pH 6.0, pH 6.0 + H2O2) represented as percent of initial DLS count rate. [H2O2] = 0.25 mM, T = 37°C. (E) Fluorescence microscopic images of the nanoprobes before (left) and after (right) activation. [H2O2] = 0.25 mM, pH = 6, incubation time: 1 h. (F) Silenced (‘OFF’ state, green trace) and activated (‘ON’ state, red trace) nanoprobes possess different emission maxima. (G) H2O2-triggered (0 - 1 mM) fluorescence signal acquired using a single filter set (λex = 745 ± 15 nm, λem = 800 ± 10 nm) consists of a mixture of both ‘OFF’ and ‘ON’ state emission (raw signal, yellow). The NPs' distinct spectral fingerprints allow quantitative decomposition of the mixed raw signal into its individually resolved fluorescence constituents, i.e., ‘OFF’ state (green) and ‘ON’ state signal (red), using multispectral acquisition (λex = 745 ± 15 nm, emission scanned from 790 to 850 nm) and software-based spectral unmixing algorithms. Higher concentrations of H2O2 yield faster and greater fluorescence activation.
