Figure 1.
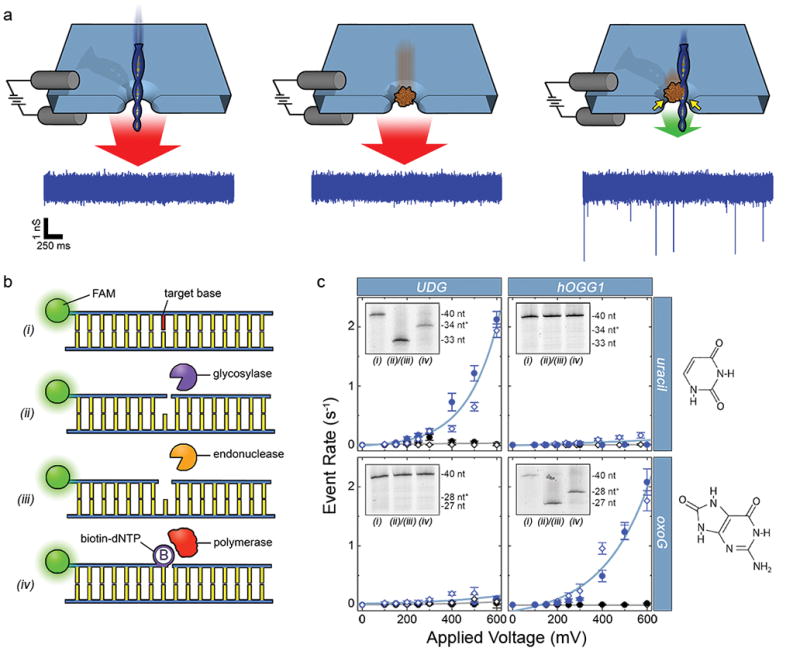
(a) Depiction of the selective SS-nanopore assay. Individual passage of a short DNA (left) or a chaperone protein (MS, center) yields no events due to the high translocation speed (red arrows); a DNA-protein complex (right) interacts with the pore walls (yellow arrows), resulting in slower translocation speed (green arrow) and resolvable events. Sample conductance traces at bottom were measured at 300 mV using 75 bp DNA (500 nM) with a synthetic biotin. (b) Schematic representation of the general labeling approach. (i) A duplex DNA molecule featuring a target base element (red). (ii) A glycosylase recognizes and excises the base element (diagram shows activity of a bifunctional glycosylase that nicks the phosphate backbone 3′ to the excision). (iii) An AP endonuclease cuts the backbone 5′ to the excision. (iv) A gap-filling polymerase incorporates a single biotinylated nucleotide at the modification position. (c) SS-nanopore analyses of 250 nM DNA oligonucleotides featuring either a single uracil (at nucleotide position 34, top) or a single oxoG (at nucleotide position 28, bottom). Data points indicate measurements on treated DNA with (blue) and without (black) MS. Filled circles and open diamonds are independent measurements on different SS-nanopore devices and all lines are exponential fits to the data. Dramatic increases in event rate are measured for DNA-MS when a glycosylase specific for the target base is used (blue data, upper left and lower right). Almost no effect is observed for mismatched glycosylase (blue data, upper right and lower left). Insets: denaturing gel analyses of the same DNA constructs (steps numbered as in (b)). Lane 1: annealed oligonucleotide; lane 2: following glycosylase/endonuclease treatment; lane 3: following T4(exo-) fill-in. * indicates DNA length plus biotin tag. Right: molecular structures of the target bases.
