Figure 1. Experimental Results.
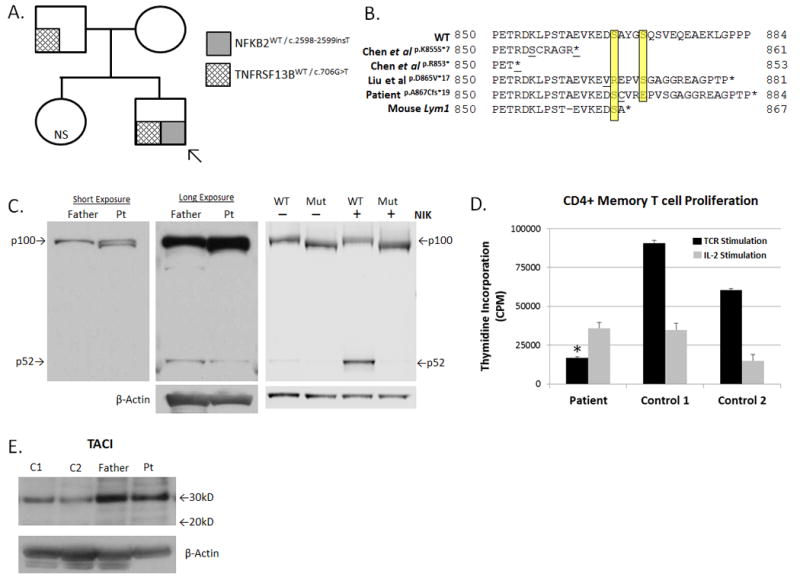
(A). Pedigree and Mutational Analysis. Arrow indicates proband (“patient”).
(B). Predicted NFκB2 Protein Sequence Alignment. Yellow highlighting denotes Ser866 and Ser870. Underlined amino acids indicate sites of the first divergent codon.
(C). NFκB2 Protein Analysis: (Left & Middle panels) Fresh PBMC-derived protein lysates from the patient (pt) and his father were probed with an anti-NFKB2 antibody (Cell Signaling #4882). Note “double” p100 band and a weak p52 band in the patient lane. (Right Panel): Lysates from NFKB2-transfected HEK293 cells, with or without NIK co-transfection, followed by NFKB2 western blot. In all panels, protein loading was normalized by BCA quantification and β-actin immunoblot.
(D). TCR-Mediated Proliferation: Negatively selected, untouched CD4+ memory T cells were isolated from fresh PBMCs (Miltenyi Biotec). Cells were stimulated with an anti-CD3 antibody (1μg/mL, Biolegend, Clone OKT3, black bars) or human IL-2 (2000 IU/mL, gray bars) for 120 hrs, and proliferation was quantified by tritiated thymidine incorporation performed in triplicate. Data are given as mean ±SEM. *p < 0.05 for patient versus controls (individually and mean).
(E). TACI Protein Analysis: Fresh PBMC-derived protein lysates from the patient, his father, and two unrelated controls (C1, C2) were probed with an anti-TACI antibody (Sigma, PRS2395). The wild-type TACI protein yields a band of ~32 kD, while the p.E236* truncated TACI protein is predicted to migrate at ~26 kD. No truncated TACI protein bands are observed. Protein loading was normalized by BCA quantification & β-actin immunoblot.
Abbreviations: NS, not sequenced; MW, molecular weight; CPM, counts per minute.
