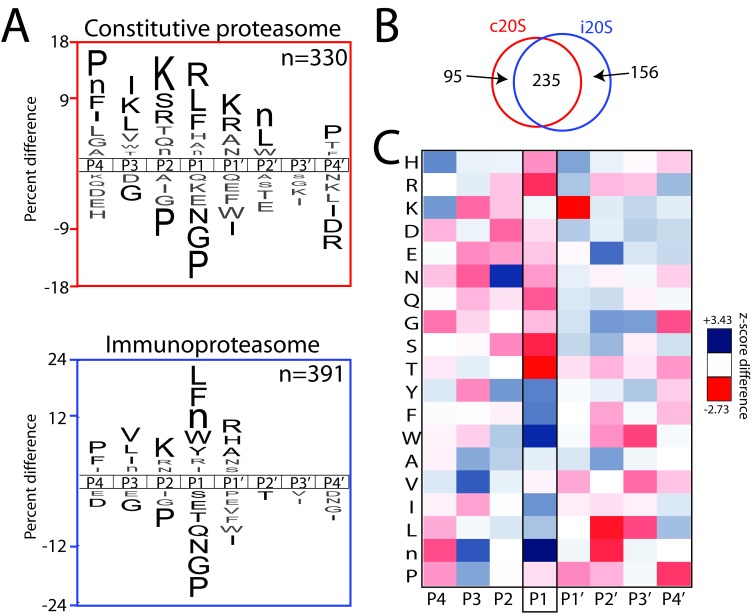
Figure 1. Global substrate specificity profiling of the iP and cP with Multiplex Substrate Profiling by Mass Spectrometry (MSP-MS) reveals shared and differential substrate specificity features.
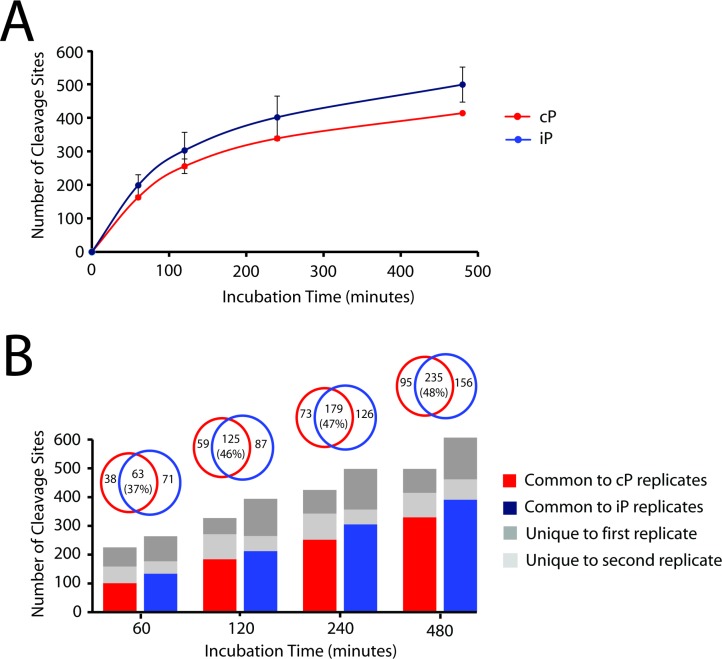
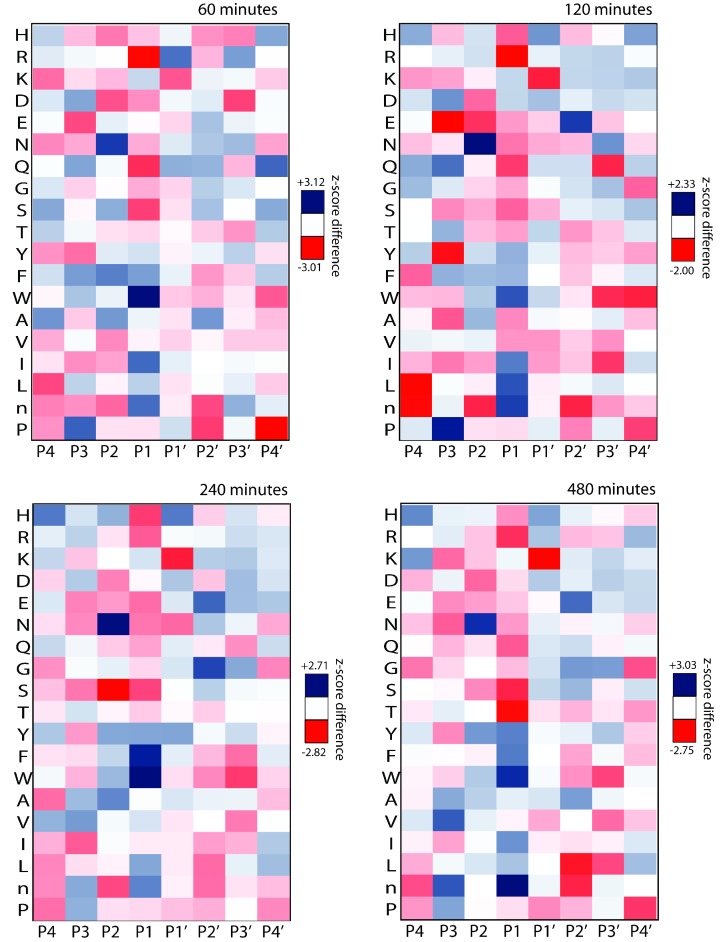
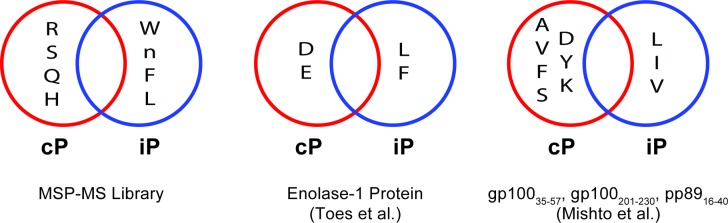
(A) iceLogo representations of iP and cP substrate specificity (P4–P4ʹ) at the 480 min assay time point (p≤0.05 for non-grayed residues (Colaert et al., 2009); ‘n’ is norleucine). (B) Quantification of the total shared and non-overlapping iP- and cP-derived cleavages in the peptide library at the 480 min assay time point. Venn diagrams for additional assay time points are provided in Figure 1—figure supplement 1, demonstrating a time-dependent increase in cleavage overlap. (C) Heat map representation of iP and cP specificity differences using Z-scores (Colaert et al., 2009) calculated for the P4-P4ʹ positions. Differences in P1 specificity are highlighted. Heat maps for additional time points are provided in Figure 1—figure supplement 3. MSP-MS revealed that the iP has an increased preference for certain bulky, hydrophobic amino acid residues at the P1 position, whereas the cP has an increased preference for smaller and polar amino acid residues at the P1 position. A qualitative comparison of the P1 specificity differences identified using the MSP-MS library with those reported in Toes et al., 2001 and Mishto et al., 2014 is provided in Figure 1—figure supplement 4. Two biological replicates were assayed for each proteasome and only overlapping cleavages between replicates are reported. An analysis of biological (Figure 1—figure supplement 1) and technical (Supplementary file 1) reproducibility is provided.