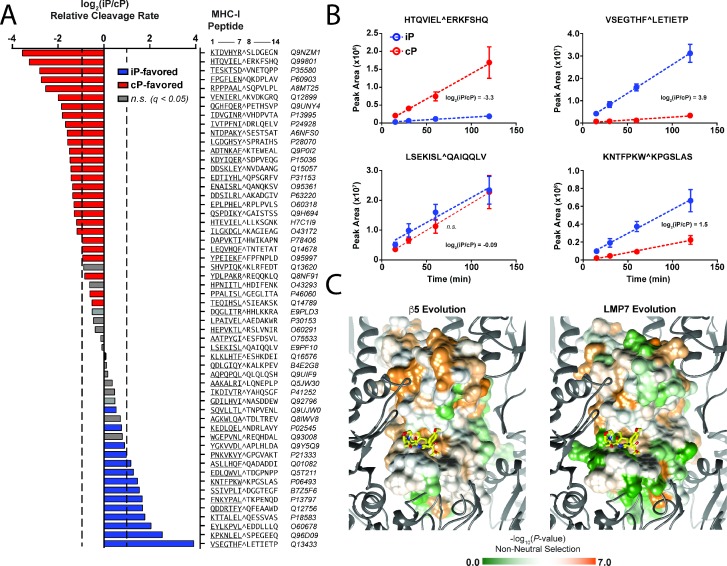
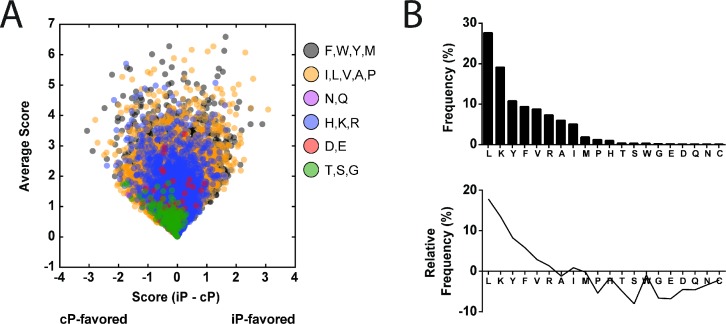
Figure 4. Differential substrate specificity of the iP alters MHC I peptide cleavage and is likely due to neutral evolution from the ancestral cP.
(A) Label-free quantitation of relative iP and cP cleavage rates against a library of synthetic peptides derived from sequences flanking sites of MHC I peptide processing (Bassani-Sternberg et al., 2015). Residues corresponding to the C-terminal portion of a given MHC I peptide are at positions 1–7 in the peptide library, whereas residues corresponding to the subsequent parent protein sequence are at positions 8–14. Average relative cleavage rates (log2) are provided for iP and cP substrates between positions 7 and 8 (P1/P1ʹ) for all peptides in the library that underwent cleavage at this site and for which quantification in all replicates was possible (n = 4). Two-fold differences in relative cleavage rate (log2 = 1) are indicated with a dashed line. Non-grayed bars represent statistically significant (q < 0.05) differences in selectivity as determined using a Student’s t-test (see Statistical methods). (B) Example kinetic traces from the MHC I peptide library time course showing cleavages following hydrophobic residues that have either high selectivity or no significant selectivity for the iP. Mean peak areas are reported with error bars representing the standard deviation (n = 4). (C) Evolutionary selection of residues in the β5 and LMP7 subunits across species that contain both proteasome isoforms. The β5 subunit has undergone more significant non-neutral evolution (residues are colored in orange) compared to the LMP7 subunit. Differences in evolutionary selection may account for the divergence in LMP7 and β5 cleavage specificity. Sequence alignments are provided in Figure 4—figure supplement 2.