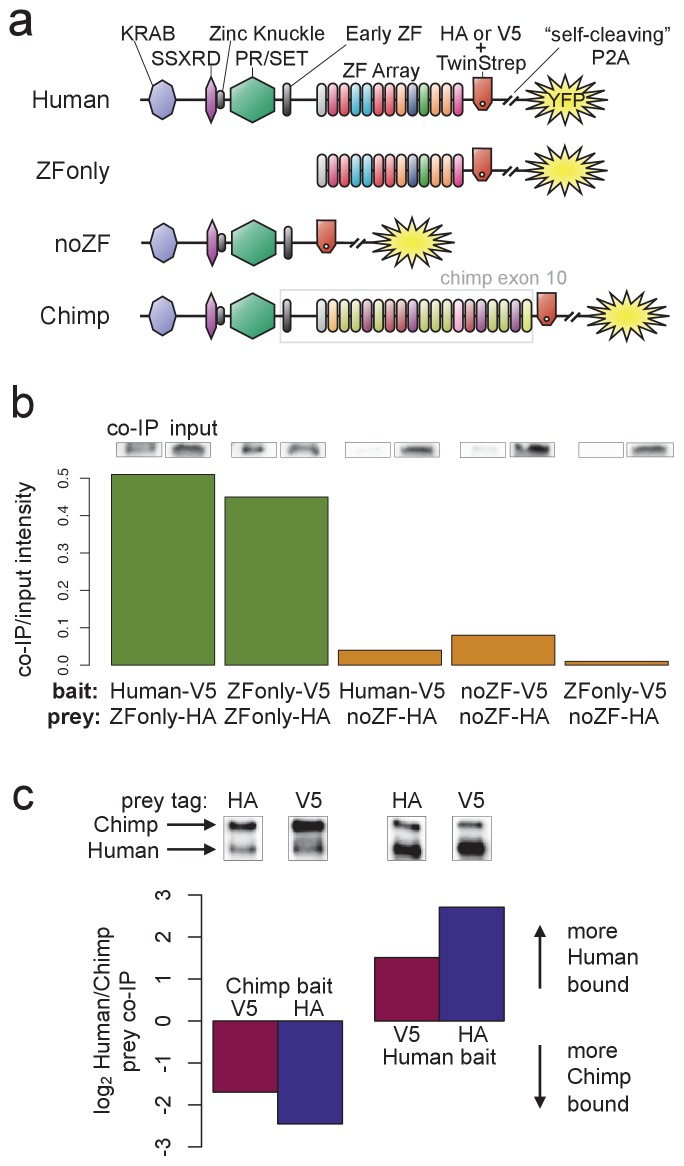
Figure 5. PRDM9 multimer formation is mediated by the ZF domain in an allele-biased manner.
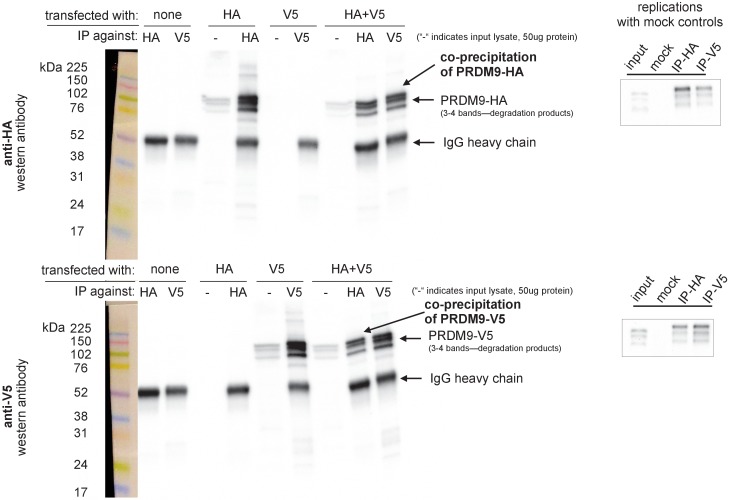
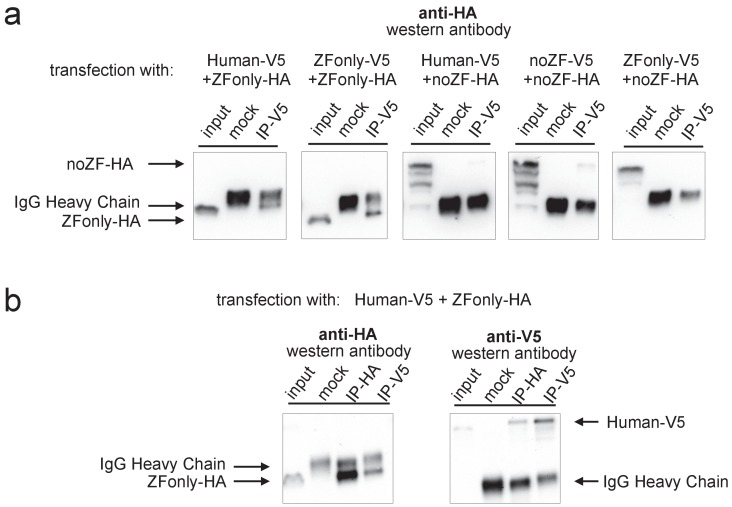
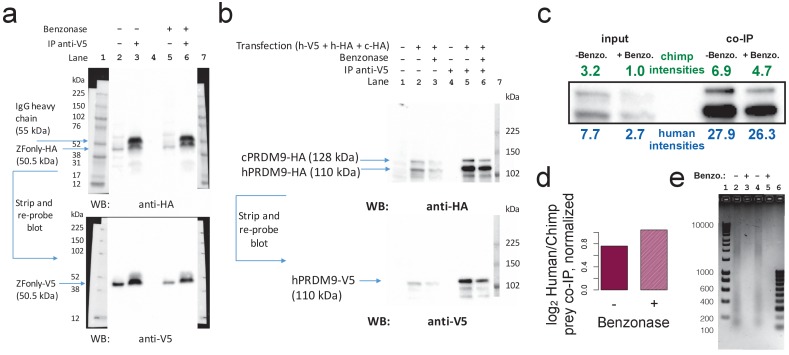
(a) Overview of the different C-terminally tagged PRDM9 constructs used. Both an HA and a V5 version of each construct were generated for co-IP experiments. (b) Barplot showing the relative intensity of western blot co-IP bands normalized to input bands (from 50-μg of total lysate protein) for each combination of bait and prey constructs. Whenever both bait and prey contain the zinc-finger domain (green bars), the co-IP signal is much stronger than when either or both constructs lack a ZF domain (orange bars). See Figure 5—figure supplements 1 and 2 for complete westerns with mock controls. (c) Barplot showing the results of competitive co-IP experiments performed in cells transfected with both Human and Chimp as prey (with the same epitope tag) and either Human or Chimp as bait (with a complementary epitope tag). Bars indicate the relative co-IP band intensity for Human and Chimp prey constructs when pulled down with either Chimp or Human bait. When Human is used as bait, more Human prey is pulled down; when Chimp is used as bait, more Chimp prey is pulled down (and this holds for both directions of HA/V5 tagging).