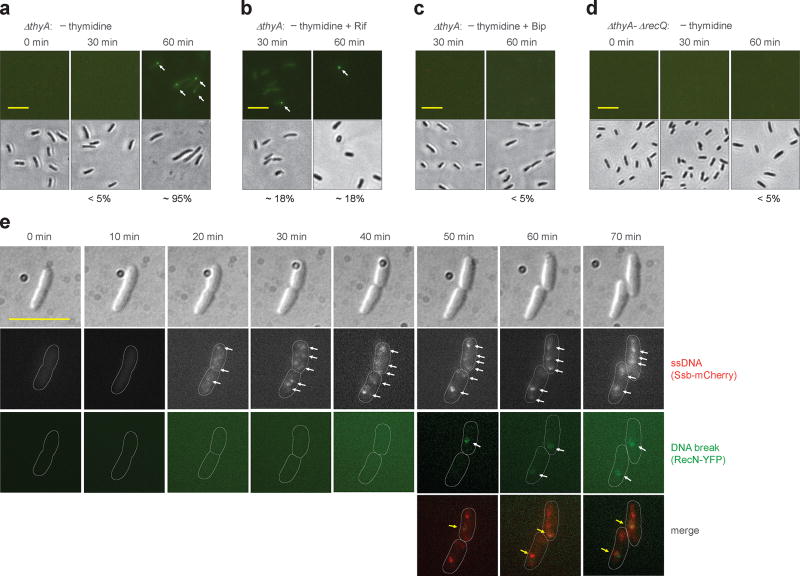
Figure 4. Double-strand DNA breaks associated with TLD.
a, Prevalence of ΔthyA cells with double-strand DNA breaks during T-starvation. Strain 4258 (recN-yfp) was pre-induced with 0.1% arabinose for 2.5 h and then starved for thymidine for the indicated times followed by examination by fluorescence (upper panels) or bright field microscopy (lower panels). Arrows indicate fluorescent foci. Scale bar (in all panels) represents 10 µm. Prevalence of cells with at least one fluorescent focus is shown as percentage below panels. b, Effect of rifampicin on presence of DNA breaks during T-starvation. Methods were as in panel a except rifampicin (50 µg ml−1) was added immediately after thymidine removal. c, Effect of 2,2’-bipyridyl (Bip) on presence of DNA breaks during T-starvation. Methods were as in panel a except for addition of 0.5 × MIC Bip immediately after thymidine removal. d, Effect of recQ on formation of DNA breaks during T-starvation. Methods were as in panel a but with strain 4262 (ΔthyA-ΔrecQ). e, Co-localization of DNA breaks and single-strand DNA regions during T-starvation. Ssb-mCherry was constitutively expressed under the native ssb promoter in the chromosome of a ΔthyA recN-yfp mutant (strain 4315). Two typical thymidine-starved cells are shown for appearance of DNA breaks in the single-strand DNA region. Arrows indicate fluorescent foci. About 60% of DNA breaks (green RecN-YFP foci, n = 30) co-localized with regions of single-strand DNA. All images are typical from pictures obtained from 3 independent experiments. For panels a-d, 200 cells for each group were randomly selected for analysis of generation of DNA breaks. Detection of double-strand DNA breaks by E. coli RecN was verified by finding the same results with Bacillus subtilis RecN expressed in E. coli (see Supplementary Fig. 9).