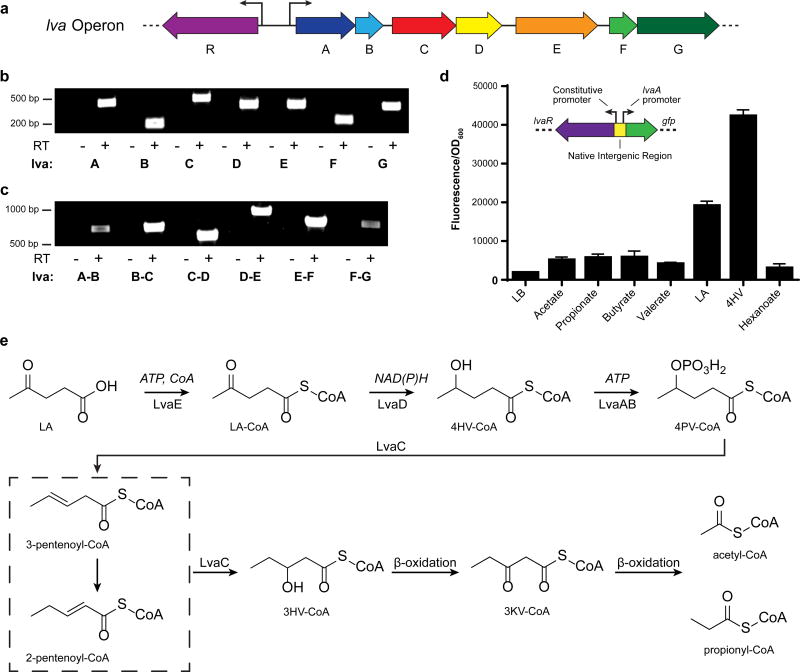
Figure 1. Genetic characterization and proposed catabolic activity of the P. putida lva operon.
a, Organization of the lvaRABCDEFG (9,323 bp) operon. b, Reverse Transcriptase (RT) PCR demonstrates that each gene is expressed in cells grown on LA. Samples were compared with the negative control (-RT) where reverse transcriptase was omitted from the reaction (n=1). c, RT-PCR of cDNA created with primer JMR237 demonstrates that the operon is polycistronic. Note that a product spanning each intergenic region was observed (n=1). d, lva operon induction assay. GFP fluorescence was measured from LB-cultures supplemented with various organic acids (20 mM) (n=3, biological). Error bars represent s.d. Insert shows the schematic of transcriptional GFP fusion used to test induction of the lva operon. lvaR was cloned onto a plasmid containing its native constitutive promoter and the native promoter region for lvaA. The fluorescent protein sfGFP was cloned in place of lvaA. e, Proposed pathway for LA metabolism. LA, levulinic acid; 4HV, 4-hydroxyvalerate; 3HV, 3-hydroxyvalerate; LA-CoA, levulinyl-CoA; 4HV-CoA, 4-hydroxyvaleryl-CoA; CoA, coenzyme-A; ATP, adenosine triphosphate; 4PV-CoA, 4-phosphovaleryl-CoA; 3KV-CoA, 3-ketovaleryl-CoA; NAD(P)H, Nicotinamide adenine dinucleotide (phosphate) reduced; GFP, green fluorescent protein.