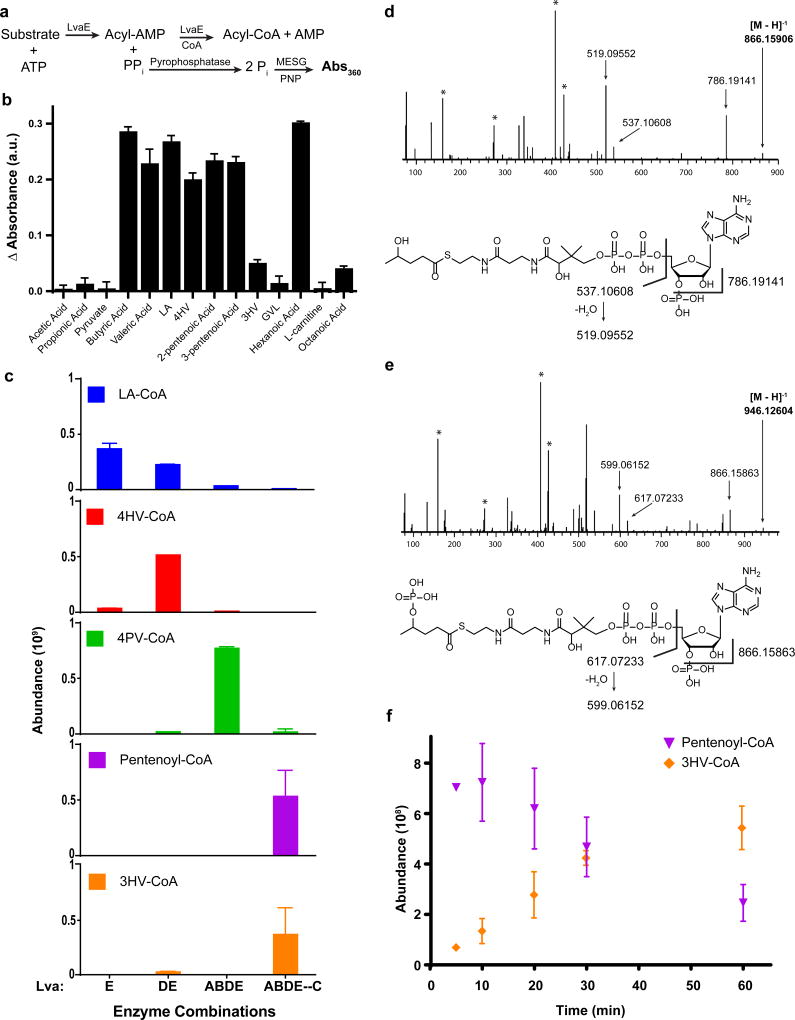
Figure 2. Enzymatic activity and pathway characterization for lva operon.
a, CoA-ligase activity assay schematic. Using the Enzchek® Pyrophosphatase Assay kit, the amount of pyrophosphate released during the CoA ligase reaction was measured as an increase of absorbance at 360 nm. b, Activity of LvaE towards short and medium chain acids (n=3, technical). Baseline subtraction was performed on all samples with a control reaction containing no substrate, indicated by Δ absorbance. c, CoA species abundance in LC/MS analysis of in vitro enzyme combinations following 30 min incubation. Reactions contained LA, CoA, ATP, and NAD(P)H with varying enzyme combinations (n=3, technical). ABDE—C indicates that the LvaABDE reaction was performed first, metabolites were separated from LvaABDE, and the resulting solution was supplemented with LvaC. The reaction confirms that LvaC is capable of converting 4PV-CoA to 3HV-CoA. d, MS/MS spectra for 4HV-CoA. Assignment of selected fragments from 4HV-CoA below. e, MS/MS spectra for 4PV-CoA. Assignments of selected fragments from 4PV-CoA below. The masses between the selected fragments of 4PV-CoA and 4HV-CoA differ by the mass of PO3H (79.967), indicating 4PV-CoA contains a phosphate group not found in 4HV-COA. Bold values indicate the mass of the parent ion. Peaks identified with the symbol (*) are fragments resulting from coenzyme A. See Supplementary Figure 6 and Supplementary Table 2 for additional fragmentation information. f, Abundance of pentenoyl-CoA and 3HV-CoA over a 60 min timecourse for a mixture of LvaABCDE, LA, CoA, ATP, and NAD(P)H (n=3, technical). AMP, adenosine monophosphate; PPi, pyrophosphate; Pi, phosphate; MESG, 2-amino-6-mercapto-7-methylpurine ribonucleoside; PNP, purine nucleoside phosphorylase; Abs, absorbance; a.u., arbitrary units. Error bars represent s.d.