FIGURE 5.
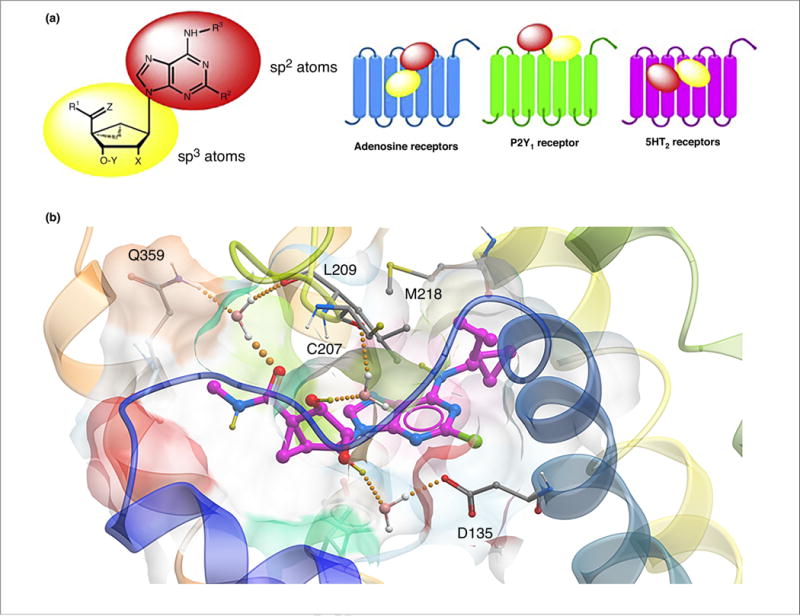
(a) Separate roles of the two moieties in rigid nucleosides in recognition at ARs and other sites, as illustrated for the general case of (N)-methanocarba adenosine derivatives. The relative orientations in the binding sites of ARs (based on X-ray structures of other nucleosides bound in the A2AAR), P2Y1R (from X-ray structure of the MRS2500 (3) complex) and in the 5HT2BR, as predicted following induced fit docking and molecular dynamics simulations, are contrasted. Typical substituents: X = H, phosphate; Y = H, OH; Z = H2, O; R1, R2, R3 = H, alkyl, O-alkyl; ethynyl, etc. (b) Hypothetical binding mode of antagonist MRS7185 (15) at the h5HT2BR. The H-bonding contacts (all through water bridges) are: 5′-carbonyl with L209 backbone (EL2) and Q359 (7.32), the 2′-hydroxyl to D135 (3.32) and the 3′-hydroxyl to C207 backbone (EL2). M218 (5.39) forms a hydrophobic contact with the N6 group.
