Table 1.
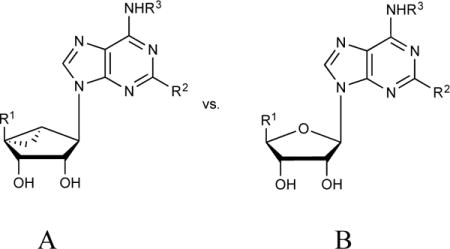
Interactions of representative adenosine derivatives with receptors and transporters. This simplified table indicates the typical contributions of each structural feature to the overall affinity of these nucleosides at the indicated target, with the beneficial gain in binding affinity indicated as +++ > ++ > + > −. Values were determined from several examples and are not necessarily general to all examples.
| ||||||
|---|---|---|---|---|---|---|
| Structural feature | hA1AR | hA3AR | mA3AR | h5HT2B/2C | hDAT | hNET |
| References: | 62, 35, 72 | 62, 35, 72 | 62 | 58, 62 | 35, 72 | 35, 72 |
| Ribose modifications (A vs. B), R1 at 5′ | ||||||
| A (N)-methanocarba | +++ | +++ | +++ | +++ | +++ | + |
| 5′-CONHCH3 | +++ | +++ | +++ | ++ | +++ | + |
| 5′-COOMe or 5′-COOEt | ++ | ++a | − | ++ | +++ | + |
| 5′-COOPr | ND | ++ | + | + | +++ | − |
| 5′-COO-i-Pr | ND | + | − | + (2C) | − | − |
| 5′-COOH | ND | + | − | + | + | − |
| 5′-CONH-(CH2)2-NH2 | − | − | − | + | + | − |
| 4′-truncated | + | ++a | + | + | − | ND |
| B 9-riboside (CH2OH) | +++ | ++ | + | ++ | ++ | + |
| Adenine modifications: R2 at C2; R3 at N6 | ||||||
| C2-Cl | +++ | +++ | +++ | ++ | − | − |
| C2-C≡C-(5-Cl-thien-2-yl) | − | +++ | ++ | + | +++ | + |
| N6-Me | ++ | +++ | ++ | + | +++ | + |
| N6-Pr | ++ | +++ | ++ | ND | ++ | − |
| N6-CH(cPr)2 | +++ | ++ | ND | +++ | − | − |
| N6-CH2Ph | ++ | +++ | +++ | ++ | − | − |
| N6-(Me)2 | − | + | + | ++ | − | − |
| 1-deaza | +++ | +++ | ++ | + | − | − |
ND, not determined.
reduces agonist efficacy.
