Figure 2.
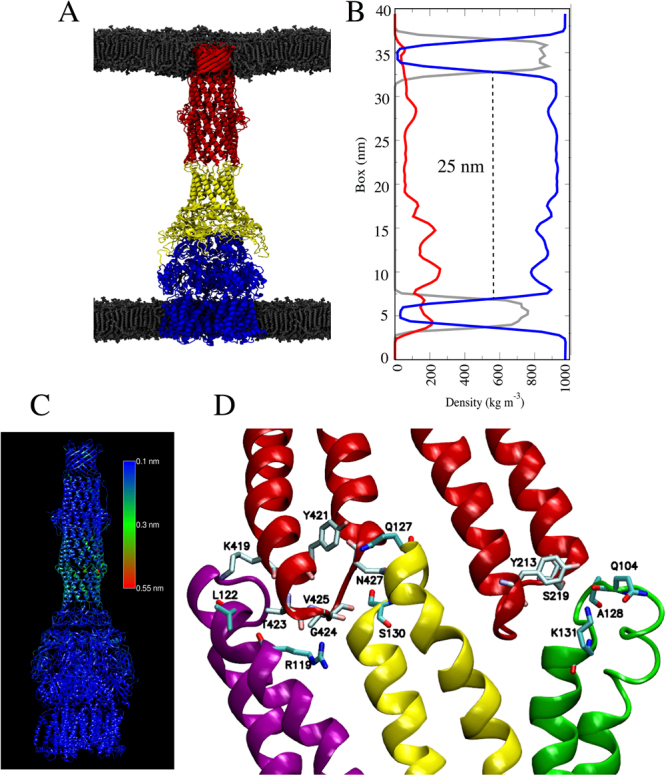
Molecular dynamics simulation of the fully assembled MexAB-OprM efflux pump. (A) The tripartite pump (red: OprM, yellow: MexA, blue: MexB) was embedded in two POPC bilayers (gray lipids) to mimic the double membrane bilayers of Gram-negative bacteria. For clarity, water molecules are not shown. (B) Mass density profile of the simulated system highlighting the peaks corresponding to the protein structure (red), the POPC lipids (gray), and the water molecules (blue). The density highlights the thickness of the periplasmic region as denoted by the distance between the phosphate groups of the corresponding inner leaflets (25 nm). (C) Root mean square fluctuations (RMSF) of the backbone atoms of the whole pump, highlighting higher fluctuations in the contact region between the periplasmic and the outer membrane domains. Fluctuations mostly range between 0.2 and 0.4 nm. (D) Close-up of the residues interacting between the periplasmic adaptor and outer membrane proteins during the MD simulation. Note that a single OprM monomer (red ribbons) is able to interact with three periplasmic protomers (green, yellow, and purple ribbons).
