FIG 1 .
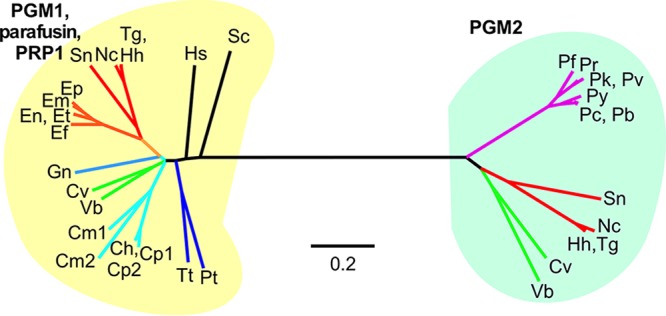
Phylogeny of PGMs in the Apicomplexa and their relatives. Unrooted tree based on alignment of PGM1 and PGM2 sequences from the following organisms: Toxoplasma gondii (Tg), Neospora canimun (Nc), Hammondia hammondi (Hh), Sarcocystis neurona (Sn), Eimeria maxima (Em), Eimeria praecox (Ep), Eimeria falciformis (Ef), Eimeria tenella (Et), Eimeria necatrix (En), Eimeria brunetti (Eb), Plasmodium falciparum (Pf), Plasmodium chabaudi (Pc), Plasmodium berghei (Pg), Plasmodium yoelii (Py), Plasmodium knowlesi (Pk), Plasmodium vivax (Pv), Gregarina niphandrodes (Gn), Cryptosporidium parvum (Cp), Cryptosporidium muris (Cm), Cryptosporidium hominis (Ch), Chromera velia (Cv), Vitrella brassicaformis (Vb), Paramecium tetraurelia (Pt), Tetrahymena thermophila (Tt), Saccharomyces cerevisiae (Sc), and Homo sapiens (Hs). The sequences (accession numbers are shown in brackets) were Toxoplasma gondii PGM1 (TgPGM1) [TGME49_285980] and TgPGM2 [TGME49_318580], Neospora canimun A0A0F7UAD1 PGM1 (NcPGM1) [NCLIV_014450 has wrong first exon] and NcPGM2 [NCLIV_010960], Hammondia hammondi PGM1 (HhPGM1) [HHA_285980] and HhGPM2 [HHA_318580], Sarcocystis neurona PGM1 (SnPGM1) [SN3_00900660] and SnPGM2 [SN3_01700255], Eimeria maxima PGM1 (EmPGM1) [EMWEY_00024400], Eimeria praecox PGM2 [EPH_0036200], Eimeria falciformis PGM1 (EfPGM1) [EfaB_MINUS_40637.g2669_1], Eimeria tenella PGM1 (EtPGM1) [ETH_00002785], Eimeria necatrix PGM1 (EnPGM1) [ENH_00082780], Eimeria brunetti PGM1 (EbPGM1) [EBH_0076610], Plasmodium falciparum GGM2 (PfGGM2) [PF3D7_1012500], Plasmodium reichenowi GGM2 (PrGGM2) [PRCDC_1011900], Plasmodium chabaudi PGM2 (PcPGM2) [PCHAS_1211600], Plasmodium berghei PGM2 (PbPGM2) [PBANKA_1210900], Plasmodium yoelii PGM2 (PyPGM2) [PY17X_1214100], Plasmodium knowlesi PGM2 (PkPGM2) [PKNH_0812300], Plasmodium vivax PGM2 (PvPGM2) [PVX_094845], Gregarina niphandrodes PGM1 (GnPGM1) [GNI_111250], Cryptosporidium parvum PGM1.1 (CpPGM1.1) [cgd2_3270] and CpPGM1.2 [cgd2_3260], Cryptosporidium muris PGM1.1 (CmPGM1.1) [CMU_003300] and CmPGM1.2 [CMU_003310], Cryptosporidium hominis PGM1 (ChPGM1) [Chro.20343], Chromera velia PGM1 (CvPGM1) [Cvel_11076] and CvPGM2 [Cvel_22350], Vitrella brassicaformis PGM1 (VbPGM1) [Vbra_3443] and VbPGM2 [Vbra_19870], Paramecium tetraurelia GPGM1 (PtGPGM1) [AAB05649.2], Tetrahymena thermophile PGM2 (TtPGM1) [AAB97159.1], Saccharomyces cerevisiae PGM1 (ScPGM1) [CAA89741.1], and Homo sapiens PGM1 (HsPGM1) [AAA60080.1]. Branch colors reflect species relationships with the cyst-forming coccidia in red, non-cyst-forming coccidia in orange, Plasmodium spp. in purple, Cryptosporidium spp. in light blue, gregarines in medium blue, ciliates in dark blue, and the chromerids in green. Bar, 0.2 nucleotide substitutions per position.
