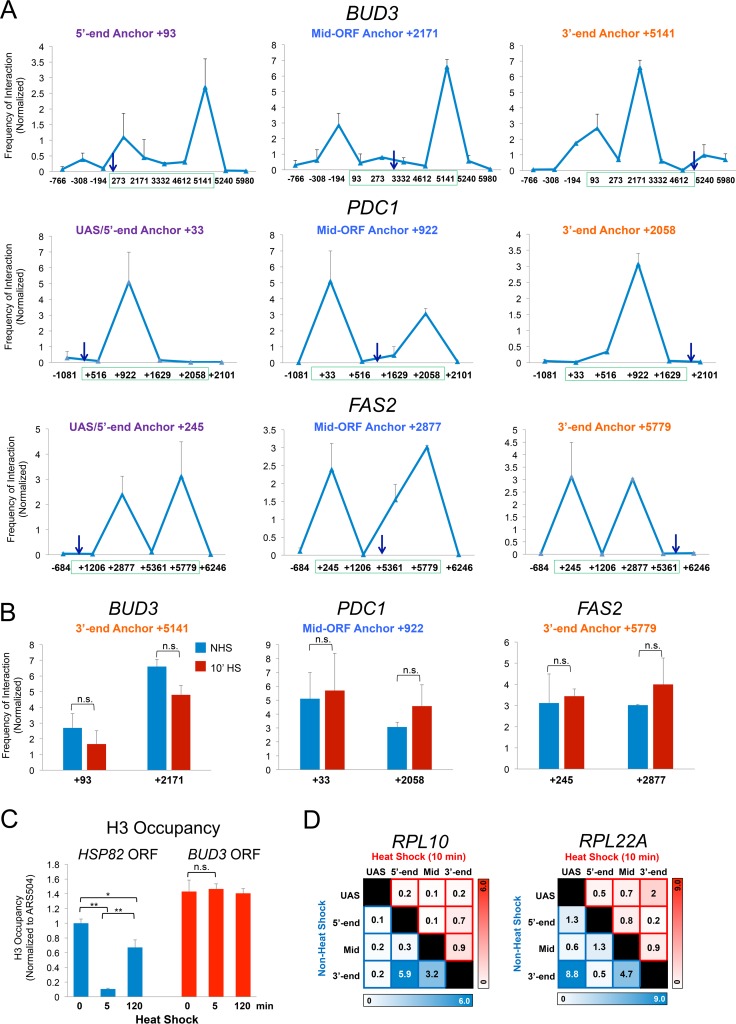
FIG 3.
TaqI-3C detects intragenic interactions within constitutively expressed genes. (A) Normalized interaction frequencies within BUD3, PDC1, and FAS2 in control cells detected using the indicated anchors (arrows). These were paired with primers abutting TaqI sites located along upstream, coding, and downstream regions. The green box spans the transcribed region of each gene. Gene regions are defined in Fig. S2A in the supplemental material. Shown are means and SD; n = 2; qPCR = 4. (B) Intragenic contact frequencies within constitutively expressed genes do not significantly change following shift to 39°C. The indicated 3C interactions within BUD3, PDC1, and FAS2 were assayed in NHS and 10-min HS cells. n.s., not significant; P = 0.1 to 0.4 (BUD3), P = 0.3 to 0.8 (PDC1), and P = 0.4 to 0.8 (FAS2). Two-tailed t test was used here and elsewhere unless specified otherwise. (C) H3 occupancy within HSP82 and BUD3 coding regions prior to or following 5 or 120 min of heat shock. H3 abundance was determined using ChIP-qPCR as described in Materials and Methods. **, P < 0.01; *, P < 0.05; n.s., P > 0.5 (one-way analysis of variance test). (D) Intragenic contact frequencies detected within ribosomal protein-coding genes RPL10 and RPL22A under NHS and 10-min-HS conditions. Representations are the same as those described in the legend to Fig. 2C.