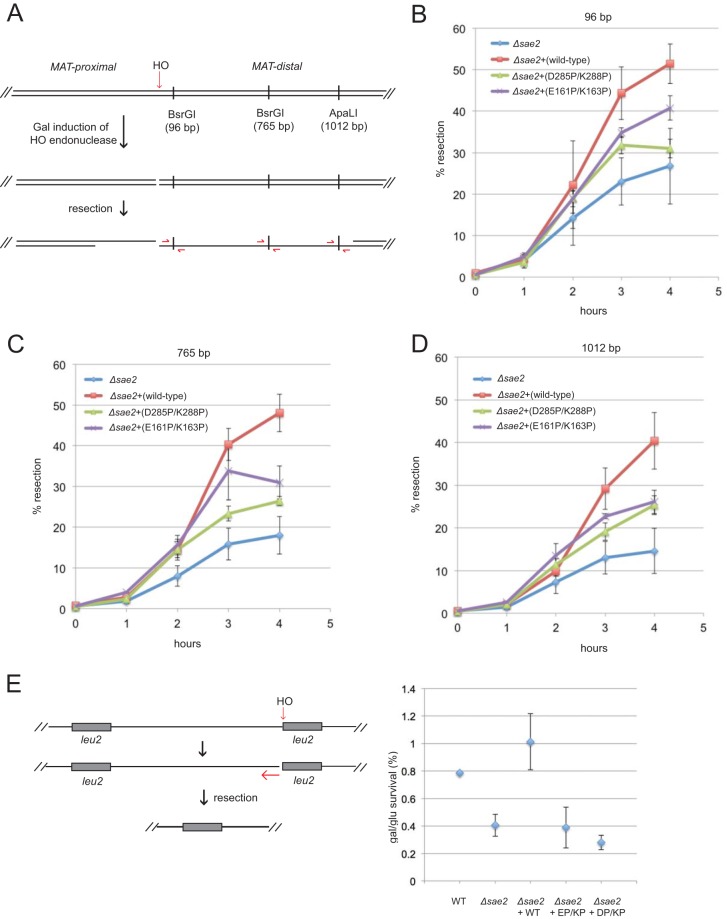
FIG 6.
Sae2 C-terminal and central domain mutations reduce resection efficiency in vivo. (A) Diagram of the MAT locus showing the site of HO endonuclease cleavage and the locations of the BsrGI and ApaLI restriction sites used to determine ssDNA levels, as shown. (B) Quantification of ssDNA at the BsrGI site located 96 bp from the HO-generated DSB in sae2Δ yeast cells expressing the vector only, wild-type Sae2, or the D285P/K288P or E161P/K163P Sae2 mutant on 2μ plasmids at various times after the addition of galactose. Error bars indicate standard errors of the means from 3 biological replicates. (C) Quantification of ssDNA at the BsrGI site located 765 bp from the HO-generated DSB in sae2Δ yeast cells as described above for panel B. (D) Quantification of ssDNA at the ApaLI site located 1,012 bp from the HO-generated DSB in sae2Δ yeast cells as described above for panel B. (E) Diagram of the HO endonuclease site adjacent one of two partial alleles of LEU2 (his4::leu2 and leu2::cs) on chromosome III. Resection from the cut site through ∼25 kb of the intervening sequence generates a LEU2 repair product by single-strand annealing (48). Wild-type or sae2Δ strains expressing the mutant alleles as indicated were plated in dilutions onto galactose, which induces HO endonuclease, or on glucose. The percentage of survivors was determined. Error bars indicate standard errors of the means from 3 biological replicates.