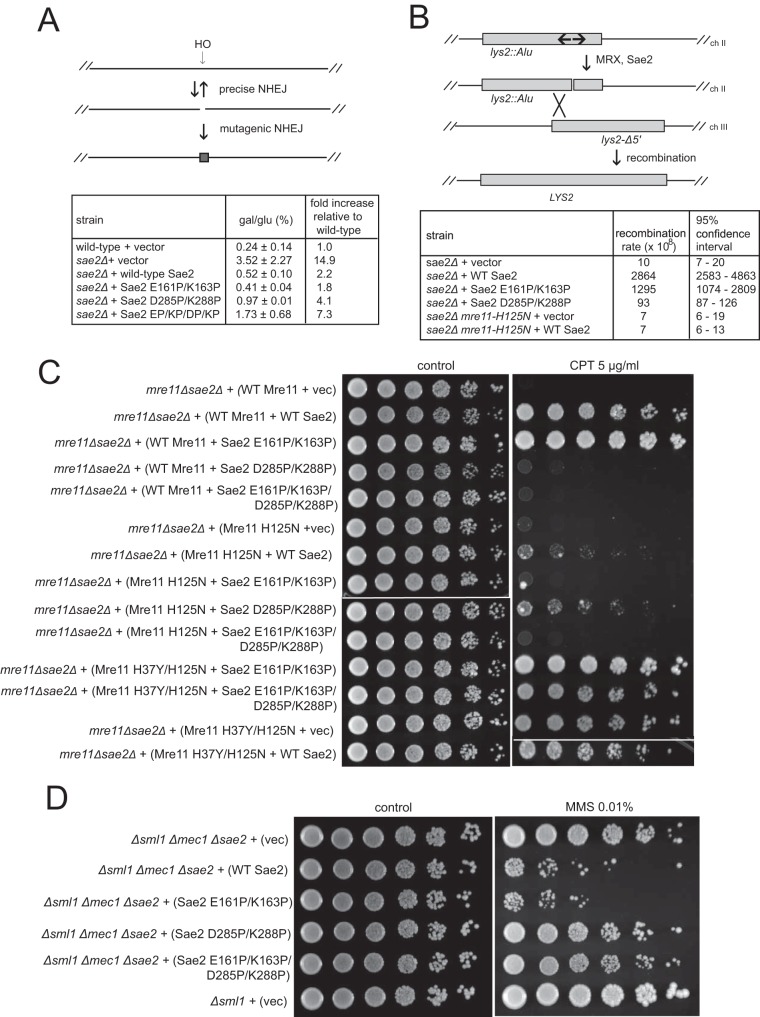
FIG 7.
Sae2 C-terminal mutations reduce Sae2 function in end joining and cruciform processing. (A, top) Diagram showing the HO endonuclease cleavage site at the MAT locus and mutagenic end joining that removes the recognition site for cleavage. (Bottom) Wild-type and sae2Δ yeast strains expressing the vector or various SAE2 alleles on 2μ plasmids, as indicated, were plated onto glucose (glu)- or galactose (gal)-containing plates, and the percentage of survivors (gal/glu) was calculated. The error shown is the standard deviation from 3 biological replicates. (B, top) Diagram showing an inverted repeat on chromosome II before and after a spontaneous DSB promoted by MRX and Sae2, as previously shown (3). Recombination between the DSB and a homologous region on chromosome III can generate a LYS2 prototroph. (Bottom) Spontaneous rates of this recombination event were measured in wild-type and sae2Δ yeast strains expressing the vector or various Sae2 alleles on CEN plasmids using fluctuation analysis. The measured rates as well as the 95% confidence intervals are shown. (C) Yeast strains deficient in MRE11 and SAE2 were complemented with FLAG-tagged sae2 alleles expressed from a 2μ plasmid under the control of the native SAE2 promoter and with yeast Mre11 expressed from the native promoter on CEN plasmids, as indicated, and were tested for CPT sensitivity as described in the legend to Fig. 2A. (D) Flag-tagged Sae2 was expressed from a CEN plasmid under the control of the native Sae2 promoter in Δsml1 Δmec1 Δsae2 yeast cells as indicated. Fivefold serial dilutions of cells expressing the indicated Sae2 alleles were plated onto nonselective medium (control) or medium containing MMS (0.01%).