ABSTRACT
The transcription factor Bach2 regulates both acquired and innate immunity at multiple steps, including antibody class switching and regulatory T cell development in activated B and T cells, respectively. However, little is known about the molecular mechanisms of Bach2 regulation in response to signaling of cytokines and antigen. We show here that mammalian target of rapamycin (mTOR) controls Bach2 along B cell differentiation with two distinct mechanisms in pre-B cells. First, mTOR complex 1 (mTORC1) inhibited accumulation of Bach2 protein in nuclei and reduced its stability. Second, mTOR complex 2 (mTORC2) inhibited FoxO1 to reduce Bach2 mRNA expression. Using expression profiling and chromatin immunoprecipitation assay, the Ccnd3 gene, encoding cyclin D3, was identified as a new direct target of Bach2. A proper cell cycle was lost at pre-B and mature B cell stages in Bach2-deficient mice. Furthermore, AZD8055, an mTOR inhibitor, increased class switch recombination in wild-type mature B cells but not in Bach2-deficient cells. These results suggest that the mTOR-Bach2 cascade regulates proper cell cycle arrest in B cells as well as immunoglobulin gene rearrangement.
KEYWORDS: B cells, cell cycle, immunoglobulin gene rearrangement, mTOR, transcription factor
INTRODUCTION
B cells play critical roles for humoral immunity by generating antibody-secreting plasma cells. B cells undergo multiple stages of discrete intermediates. During their differentiation, signaling from cell-stage-specific receptors controls developmental events, such as DNA rearrangement of immunoglobulin genes. The interleukin-7 receptor (IL-7R) is required for the proliferation and survival of B cell progenitors as well as their differentiation to pro-B cells in part by activating phosphatidylinositol-3-OH kinase (PI3K)–AKT (1). In pro-B cells, a fluctuation of IL-7R signaling controls immunoglobulin heavy (IgH) chain rearrangement to produce the pre-B cell receptor (pre-BCR), which is composed of IgH, surrogate light chain, Igα, and Igβ. Upon successful generation of pre-BCR, pro-B cells become large pre-B cells. When IL-7R signaling is attenuated in large pre-B cells, cells proceed to small pre-B cells, in which Ig light (IgL) chain rearrangement is dramatically induced by the activation of the SYK-BLNK-p38 pathway under pre-BCR signaling (2). Small pre-B cells become immature B cells upon successful rearrangement of IgL genes, and therefore expression of nonautoreactive BCR occurs, which then terminates further IgL rearrangement by a strong input into the PI3K-AKT cascade (3). The PI3K-AKT cascade further regulates responses of mature B cells. It represses class switch recombination (CSR), which produces the diversity of immunoglobulin genes in germinal center B cells (4). Thus, cell surface receptors and their downstream signaling cascades are essential for organizing differentiation along the entire B cell development. Considering that many of the cell surface receptors ultimately elicit cell responses by changing gene expression, it is critically important to understand transcription factors (TFs), the activities of which are regulated by the signaling pathways.
Among TFs required for B cell differentiation, FoxO1 has been well studied in terms of its regulation linking to signaling regulators. AKT directly phosphorylates FoxO1, resulting in its protein degradation (5). Importantly, PI3K-mTORC2-AKT mediates inactivation of FoxO1 protein by nuclear exclusion, contributing to cell proliferation in endothelial cells (6). In contrast to the negative regulation of FoxO1 by PI3K-AKT, FoxO1 phosphorylation by p38 kinase promotes the transcriptional activity of FoxO1 (7). The attenuation of IL-7R signaling in pro-B and pre-B cells activates FoxO1, resulting in the induction of Rag1/2 mRNA, encoding enzymes required for the rearrangement of IgH and IgL genes (2, 8, 9). Other than FoxO1, little is known about TFs in B cells that are regulated by the PI3K-AKT pathway and control cell proliferation and/or differentiation.
The transcription factor Bach2 is required for efficient commitment of common lymphoid progenitor cells to the B cell lineage as well as CSR and somatic hypermutation of Ig genes in mature B cells. Bach2 promotes lymphoid lineage differentiation by repressing myeloid cell-related genes at the divergence of myeloid and lymphoid progenitors (10, 11). After being committed to lymphoid cells, Bach2 plays essential roles for differentiation of both T- and B-lymphoid cells. In T-lymphoid cells, Bach2 controls the differentiation into CD4- and CD8-positive effector lymphocytes (12, 13). In B-lymphoid differentiation, Bach2 is required for germinal center (GC) formation, CSR, and somatic hypermutation (14). Bach2 also inhibits plasma cell (PC) differentiation by repressing Prdm1 expression (15, 16). These observations raise the possibility that activation and/or inactivation of surface receptors modulates Bach2 to alter gene expression and therefore the responses of B cells at these various differentiation stages. Supporting this possibility, we have recently revealed that the PI3K-AKT-mammalian target of rapamycin (mTOR) cascade phosphorylates Bach2 to reduce its activity. Among multiple phosphorylation sites of Bach2, phosphorylation at serine 535 (S535) prevents the nuclear localization of Bach2 protein (17). In addition, Bach2 has been shown to regulate the expressions of Rag1 and Rag2, which are repressed by PI3K-AKT signaling in the context of the pre-BCR checkpoint (2, 18). These observations suggest that signaling-mediated regulation of Bach2 function might be used in both early B cell and PC differentiation. However, the molecular mechanism and significance of receptor-mediated regulation of Bach2 are still unclear. This study reports that signaling from stage-specific receptors of B cells (i.e., IL-7R and BCR) controls Bach2 at both transcript and protein levels, regulating immunoglobulin gene recombination in both early and late stages of B cell differentiation. The mTOR-Bach2 cassette is suggested to coordinate cell cycle arrest and DNA recombination reactions in pre-B and mature B cells.
RESULTS
Regulation of Bach2 level under receptor signaling associates with Rag gene expression in early B cell development.
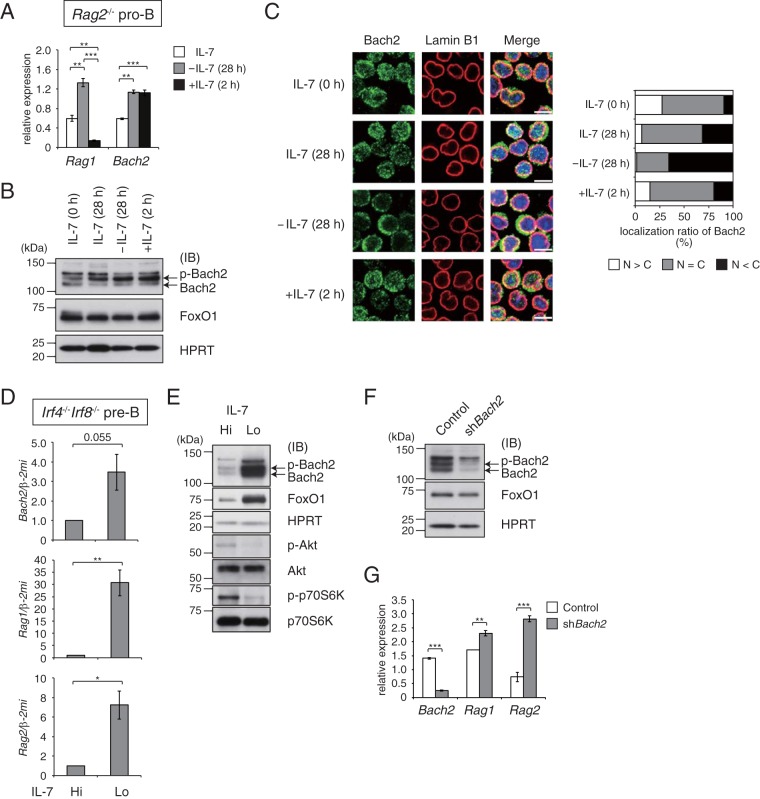
To clarify the response of Bach2 to the receptor signaling during early B cell development, we first examined the effect of IL-7R signaling upon Bach2. For this purpose, we used Rag2−/− pro-B and Irf4−/− Irf8−/− pre-B cells in which the strength of IL-7R signaling can be modified by changing the concentration of IL-7 in culture medium in the absence or presence of pre-BCR signaling (pro-B and pre-B cells, respectively) (2, 19–21). As reported previously, the withdrawal of IL-7 in Rag2−/− pro-B cells induced the expression of Rag1, and it was immediately cancelled by restimulation with IL-7 for 2 h (Fig. 1A). The expression of Bach2 was induced by IL-7 withdrawal, but it was not reduced with the restimulation with IL-7. At the protein level, IL-7 withdrawal did not affect the accumulation of Bach2, which was present mainly in a phosphorylated form, or FoxO1 (Fig. 1B). Whereas IL-7 withdrawal promoted the cytoplasmic accumulation of Bach2, restimulation of IL-7R clearly promoted the nuclear localization of Bach2 (Fig. 1C).
FIG 1.
Bach2 negatively regulates expression of Rag genes in Rag2−/− pro-B and Irf4−/− Irf8−/− pre-B cells. (A to C) IL-7 was withdrawn for 28 h (−IL-7) from cultures of Rag2−/− pro-B cells, and the cultures were restimulated for 2 h with IL-7 (+IL-7); shown are quantitative RT-PCR analysis of Rag1 and Bach2 expression (A), immunoblot analysis of Bach2 and FoxO1 (B), and immunohistochemistry for Bach2 protein (C). (C) Bach2 (green) and lamin B1 (red) distribution in cells (left); the subcellular localization of Bach2 was evaluated by classification of cells (n = 100) for each condition into three classes (right): cytoplasm dominant (N < C), nucleus and cytoplasm (N = C), and nucleus dominant (N > C). Bar, 10 μm. (D and E) Immunoblot analysis of Bach2, FoxO1, phosphorylated Akt (p-Akt), total Akt, p-p70S6K, and p70S6K (D) or quantitative RT-PCR analysis of Bach2 and Rag1/2 expression (E) in Irf4−/− Irf8−/− pre-B cells cultured with 5.0 ng/ml (Hi) or 0.1 ng/ml (Lo) of IL-7 for 48 h. (F and G) Immunoblot analysis of Bach2 and FoxO1 (F) or RT-PCR analysis of Bach2 and Rag1/2 expression (G) in Irf4−/− Irf8−/− pre-B cells transduced with a control vector (Control) or vector targeting Bach2 (shBach2). Then, cells were sorted on the basis of GFP expression and cultured under IL-7 Lo conditions for 48 h. For panels B, E, and F, HPRT served as an internal control. IB, immunoblot; p-Bach2, phosphorylated Bach2. For panels A, D, and G, relative expression was normalized by β2-microglobulin (Β2M) mRNA, and the mean expression and standard error of the mean (SEM) data were from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next examined the effect of IL-7 withdrawal in Irf4−/− Irf8−/− pre-B cells. Compared with Rag2−/− pro-B cells, the amounts of both mRNA and protein of Bach2 were markedly increased in Irf4−/− Irf8−/− pre-B cells by IL-7 withdrawal (Fig. 1D and E). Bach2 was present in phosphorylated and unphosphorylated forms. Nuclear accumulation of Bach2 was also promoted in these cells (see Fig. S1 in the supplemental material). The expressions of Rag1/2 transcripts were also induced (Fig. 1D). We should note that while mRNA expression of Bach2 was induced in pro-B and pre-B cells when IL-7 was low, the amount of protein was significantly increased only in pre-B cells. To examine the effect of Bach2 on Rag1/2 expression, we performed knockdown of Bach2 in Irf4−/− Irf8−/− pre-B cells (Fig. 1F). The expression of Rag1/2 gene showed higher levels in cells with Bach2 knockdown than in control cells (Fig. 1G). Bach2 might directly or indirectly repress Rag1/2 expression in these cells.
mTOR complexes (mTORCs) regulate Bach2 expression and protein stability via phosphorylation at S535.
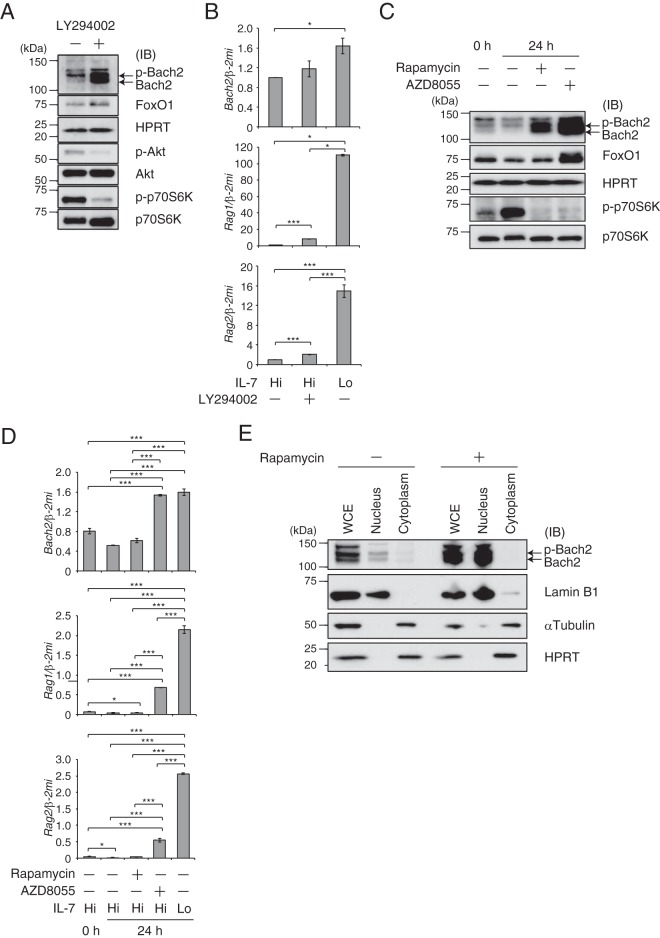
In the above-described experiments, the expression of Bach2 showed a pattern similar to that of Rag genes, which are direct targets of FoxO1. As expected, we identified FoxO1-binding peaks on the Bach2 locus by examining previously reported chromatin immunoprecipitation sequencing (ChIP-seq) data (2, 18). Binding of FoxO1 to these regulatory regions was confirmed by ChIP-PCR (see Fig. S2A and B in the supplemental material), indicating Bach2 as a direct target of FoxO1. Since IL-7R signaling is coupled to the PI3K-AKT-FoxO1 pathway, we examined the effect of the pharmacological inhibition of PI3K with LY294002 on Bach2 and FoxO1. In the presence of IL-7, the treatment with LY294002 of Irf4−/− Irf8−/− pre-B cells resulted in only a slight accumulation of FoxO1 protein (Fig. 2A). As expected from this observation, the mRNA expression of FoxO1 targets Bach2 and Rag1/2 remained low in the presence of LY294002 compared with their expression under low IL-7 conditions (Fig. 2B) (2). Surprisingly, the amount of Bach2 protein substantially increased in response to LY294002 (Fig. 2A). Nuclear accumulation of Bach2 was enhanced under such conditions (see Fig. S3 in the supplemental material). We surmise that the PI3K pathway regulates protein stability and the subcellular localization of Bach2 in pre-B cells. mTORC1 may be involved in this regulation since it directly phosphorylates Bach2 (17).
FIG 2.
Presence of two distinct signaling pathways regulating Bach2 transcripts and protein. (A and B) Immunoblot analysis (A) or quantitative RT-PCR analysis (B) in Irf4−/− Irf8−/− pre-B cells cultured in the presence of LY294002 under IL-7 Hi conditions for 48 h. (C and D) Immunoblot analysis (C) or quantitative RT-PCR analysis (D) in Irf4−/− Irf8−/− pre-B cells cultured in the presence of rapamycin or AZD under IL-7 Hi conditions for 24 h. For panels A and C, HPRT served as an internal control. IB, immunoblot; p-Bach2, phosphorylated Bach2. For panels B and D, data under IL-7 Lo conditions for 24 h are shown as a control. Relative expression was normalized by Β2M mRNA, and the expression and SEM data were from three independent experiments. *, P < 0.05; ***, P < 0.001. (E) Subcellular localization of Bach2 in Irf4−/− Irf8−/− pre-B cells cultured in the presence of rapamycin. Lamin B1 served as a control for the nuclear fraction, and α-tubulin and HPRT were used as controls for the cytoplasm fraction. WCE, whole-cell extract.
To investigate this possibility, we used rapamycin, the specific inhibitor for mTORC1, or AZD8055, the inhibitor for both mTORC1 and mTORC2, in the presence of IL-7. Treatment of the cells with rapamycin resulted in Bach2 protein accumulation without affecting the FoxO1 protein amount or inducing Bach2 mRNA (Fig. 2C and D). In contrast, AZD8055 treatment resulted in robust accumulation of Bach2 and FoxO1 proteins (Fig. 2C), with induction of both Bach2 and Rag1/2 mRNA (Fig. 2D). The treatments with these inhibitors enhanced nuclear accumulation of both Bach2 and FoxO1 (see Fig. S4A and B in the supplemental material). These findings were further verified by a biochemical fractionation of endogenous proteins in these cells, showing an increase of nuclear Bach2 in response to rapamycin (Fig. 2E). Based on these results, we suggest two distinct pathways regulating Bach2 under IL-7R. One is the direct, transcriptional regulation of Bach2 gene by FoxO1 in pre-B cells, where FoxO1 activity is negatively regulated by mTORC2 in addition to PI3K-AKT. The other is the reduction of the amount and nuclear accumulation of Bach2 protein in pre-B cells mediated by the IL-7R–PI3K–mTORC1 cascade. The above-described observations also suggest an intimate association between cytoplasmic retention and protein stability of Bach2.
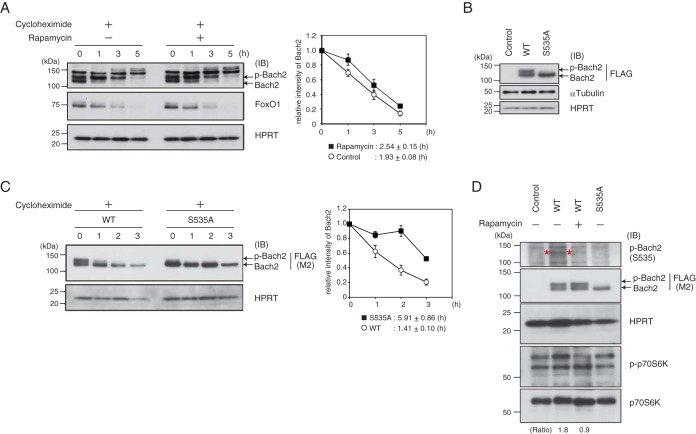
To examine whether the stability of Bach2 protein is regulated by the IL-7R–PI3K–mTORC1 cascade, the half-life of endogenous Bach2 protein in the pre-B cells in the absence of rapamycin was compared to that in the presence of rapamycin. The half-life of Bach2 was slightly increased in the presence of rapamycin, whereas that of FoxO1 was not affected (Fig. 3A). It should be noted that more of the faster-moving signal of Bach2 remained in the presence of rapamycin at 1 h after the chase than in the absence of rapamycin. To examine whether phosphorylation of Bach2 is involved in this regulation, we overexpressed wild-type Bach2 or a derivative of Bach2 with an S535A mutation, which we have reported recently as a critical phosphorylation site in mature B cells (17). Importantly, the slower-migrating band of phosphorylated Bach2 (p-Bach2) almost disappeared when Bach2S535A was expressed in these cells (Fig. 3B). Bach2S535A showed a significantly longer half-life than did wild-type Bach2 in the Irf4−/− Irf8−/− pre-B cells (Fig. 3C).
FIG 3.
mTORC1-mediated Bach2 phosphorylation at S535 promotes cytoplasm localization and protein degradation. (A) Half-life of Bach2 protein in Irf4−/− Irf8−/− pre-B cells cultured with or without rapamaycin in the presence of cycloheximide under IL-7 Hi conditions. (Left) Immunoblot analysis of Bach2 and FoxO1. (Right) Relative intensity of total Bach2 protein. The amounts of protein were normalized with HPRT. The means and SEM were from three independent experiments. (B) Immunoblot analysis in Irf4−/− Irf8−/− pre-B cells transduced with control vector or vectors expressing FLAG-tagged wild-type (WT) Bach2 or Bach2 S535A. (C) Half-life of WT Bach2 or Bach2 S535A. Irf4−/− Irf8−/− pre-B cells transduced with FLAG-tagged WT Bach2 or Bach2 S535A and cultured in the presence of cycloheximide under IL-7 Hi conditions. (Left) Immunoblot analysis of FLAG-Bach2. (Right) Relative intensity of FLAG-Bach2 protein, p-Bach2, and total Bach2. The amounts of proteins were normalized with HPRT. The means and SEM were from three independent experiments. (D) Immunoblot analysis in Irf4−/− Irf8−/− pre-B cells transduced as described for panel B and then cultured with or without rapamycin. Specific signals detected with anti-p-S535 antibody were marked with asterisks. The ratio was calculated as the division of p-Bach2 signal with anti-S535 antibody by signals detected with anti-FLAG of WT Bach2 with/without rapamycin. For panels A to D, HPRT or α-tubulin served as an internal control. Each immunoblot analysis was representative of three independent experiments. IB, immunoblot; p-Bach2, phosphorylated Bach2.
To further examine Bach2 phosphorylation, we generated a monoclonal antibody specifically recognizing Bach2 with phosphorylation of S535. Whole-cell extracts were prepared from Plat-E cells transfected with pcDNA wild-type Bach2 (WT) or Bach2 S535A, which showed two specific bands for WT Bach2 or one band for Bach2 S535A as previously reported (17). By using the monoclonal antibody, we detected two signals with WT Bach2-expressing cells (see Fig. S5 in the supplemental material). These signals were not detected with Bach2 S535A-expressing cells, suggesting that the monoclonal antibody detected Bach2 with phosphorylation of S535. We found that wild-type Bach2 overexpressed in the pre-B cells was phosphorylated at this residue, which was reduced upon the inhibition of mTORC1 (Fig. 3D). Combining these data with those of our previous report (17), we conclude that phosphorylation of Bach2 at S535 by mTORC1 promotes cytoplasmic retention of Bach2 as well as its degradation in pre-B cells.
The Bach2-Ccnd3 axis controls G1 arrest during pre-B cell development.
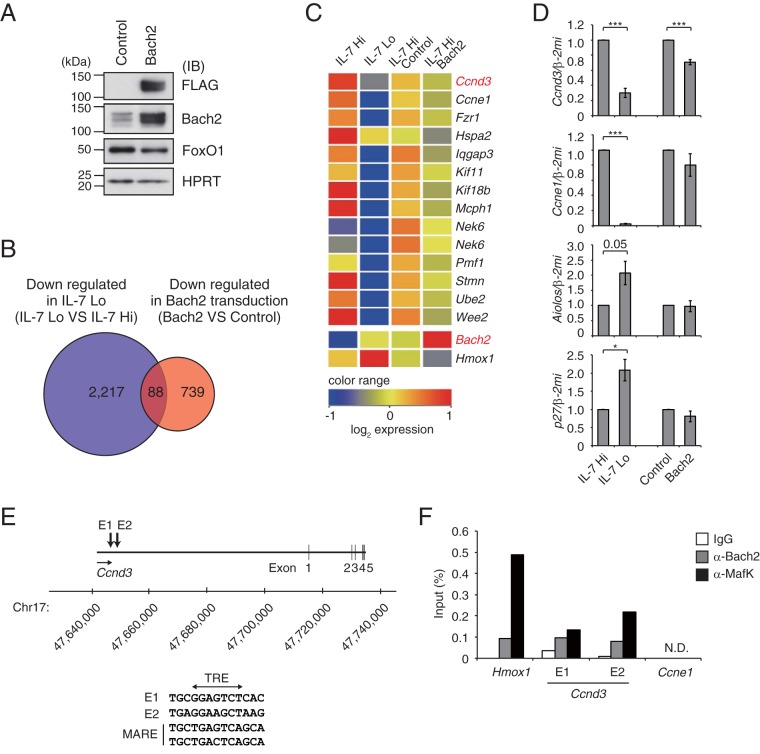
As the withdrawal of IL-7R signaling strictly regulated Bach2 expression in the pre-B cells, Bach2 might possess critical roles at this differentiation stage. To elucidate Bach2 function, we performed gene expression analysis by using Irf4−/− Irf8−/− pre-B cells transduced with control vector or Bach2 expression vector (Fig. 4A). Based on the expression of Hmox1, which is a well-known Bach2 target gene (22), we selected 827 genes downregulated more than Hmox1 (i.e., 1.3-fold) in the Bach2-transduced cells compared to the control (Fig. 4B). These data were coanalyzed with previously published data sets of genes downregulated with the withdrawal of IL-7 (2), resulting in the identification of 88 common genes (Fig. 4B; see also Table S1 in the supplemental material). Among these genes, 13 genes were related to the cell cycle progression (Fig. 4C; see also Table S2 in the supplemental material). Consistent with this, the expressions of Ccnd3 and Ccne1 were significantly repressed in Irf4−/− Irf8−/− pre-B cells with IL-7 withdrawal (Fig. 4D), and Bach2 overexpression in these cells resulted in reduced expression of Ccnd3 but not Ccne1 (Fig. 4D). The expression of Aiolos, a known repressor of Ccnd3 (23), was not altered upon Bach2 transduction. Importantly, we discovered several Maf recognition element (MARE)-like sequences, which could be bound by Bach2 and small Maf heterodimer, near the Ccnd3 locus (Fig. 4E). Bindings of both Bach2 and its partner MafK to these regions were confirmed by using quantitative ChIP-PCR (Fig. 4F). Therefore, we suggest Ccnd3 as a direct target gene of Bach2.
FIG 4.
Ccnd3 gene as a putative target of Bach2. (A) Immunoblot analysis of indicated proteins in Irf4−/− Irf8−/− pre-B cells transduced with control or Bach2 expression vector and then sorted based on GFP expression. HPRT served as an internal control. (B) Venn diagram of microarray analysis in Irf4−/− Irf8−/− pre-B cells comparing downregulated genes with IL-7 attenuation (blue) and Bach2 transduction with IL-7 Hi (red). Among 2,305 genes downregulated with IL-7 attenuation (21), 88 genes overlapped genes that were downregulated by Bach2 transduction. (C) Heatmap of cell cycle-related gene expression in a comparison of IL-7 Hi and IL-7 Lo conditions and of transduction of control vector versus Bach2. Transcripts of Hmox1 were shown to be one of the direct targets of Bach2. (D) Quantitative RT-PCR analysis of Ccnd3, Ccne1, Aiolos, and p27 in Irf4−/− Irf8−/− pre-B cells as described for panel C. Relative expression was normalized by Β2M, and the means and SEM were from three independent experiments. *, P < 0.05; ***, P < 0.001. (E) MARE-like sequences E1 and E2 at Ccnd3 locus. Bach2 binding sequences detected in these regions were shown along with MARE consensus. E, enhancer. (F) Binding of Bach2 and MafK to regulatory elements in the Hmox1, Ccnd3 E1 and E2, and Ccne1 loci in cells cultured with 0.1 ng/ml of IL-7 for 48 h, assessed by ChIP with control IgG, anti-Bach2, or anti-MafK. The data are the averages from three independent experiments. N.D., not detected.
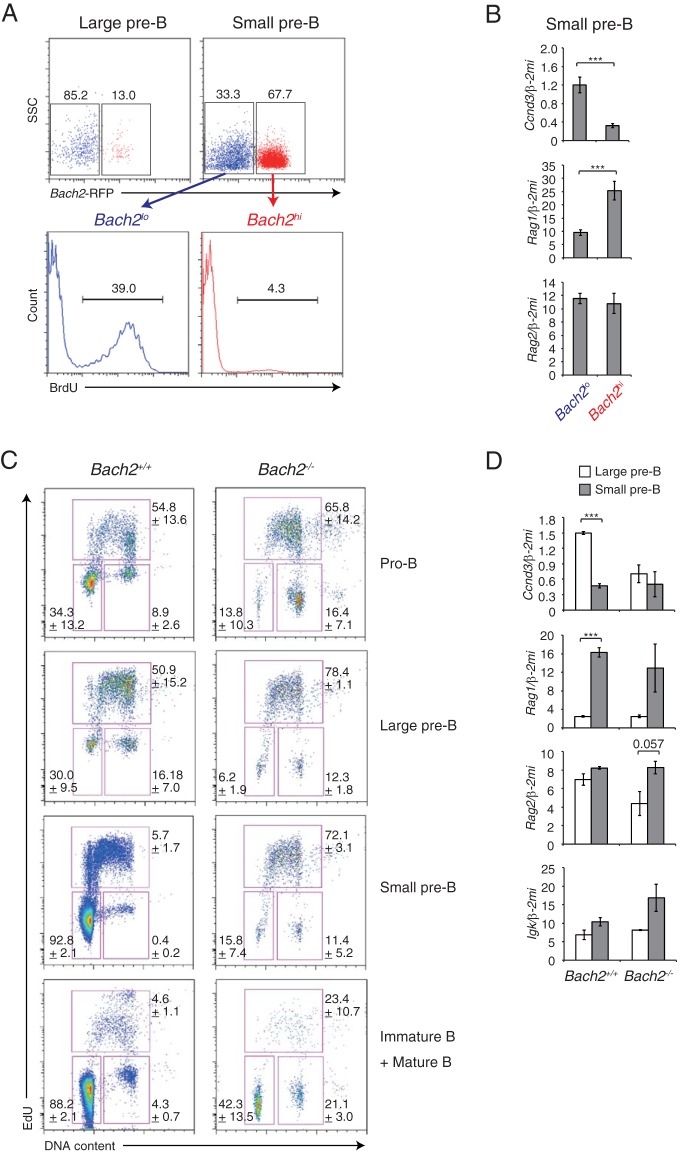
Cell cycle arrest is required for IgL rearrangement in pre-B cells (23, 24). Since an inhibition of cyclin D3 expression is expected to cause G1 arrest (24), we examined the importance of the Bach2-Ccnd3 axis for cell cycle arrest during pre-B cell differentiation. By using Bach2-RFP mice (where RFP is red fluorescent protein) (10), pre-B cells in the bone marrow were sorted into Bach2-RFP high (Bach2hi) and low (Bach2lo) fractions based on the expression of RFP (Fig. 5A). The Bach2hi population was larger in small pre-B cells than in large pre-B cells (Fig. 5A, upper panels). Intracellular staining of FoxO1 protein showed that its amount was higher in the Bach2hi population than in the Bach2lo population (see Fig. S6 in the supplemental material), confirming the regulatory relationship between FoxO1 and Bach2 gene expression. Small pre-B cells undergo cell cycle arrest (3). Surprisingly, an in vivo BrdU labeling experiment revealed that Bach2hi small pre-B cells but not Bach2lo cells underwent cell cycle arrest (Fig. 5A, lower panels). The expressions of genes in Bach2hi and Bach2lo small pre-B cells were compared. A large reduction of Ccnd3 expression was observed in the Bach2hi population compared with the Bach2lo population (Fig. 5B). Importantly, the expression of the Rag1 gene was induced in this population, whereas Rag2 expression was similar in these cells (Fig. 5B). Thus, these results suggest that Bach2hi small pre-B cells undergo cell cycle arrest and IgL rearrangement, due in part to reduced expression of Ccnd3 and increased expression of Rag1.
FIG 5.
Bach2 is required for proper G1 cell cycle arrest during B cell development. (A) Flow cytometry analysis of Bach2 transcripts in bone marrow pre-B cells of Bach2-RFP mice. Data are representative of results for four mice in one experiment. (B) Quantitative RT-PCR analysis of Ccnd3, Rag1, and Rag2 in small pre-B cell fractions sorted by RFP expression. Bach2hi and Bach2lo, Bach2-RFP high and low, respectively. (C) Cell cycle analysis by using flow cytometry in bone marrow-derived pro-B, large pre-B, small pre-B, and immature and mature B cells from wild-type (Bach2+/+) and Bach2-deficient (Bach2−/−) mice. Data are representative of results for three mice for each genotype in one experiment and are shown as means and SEM. (D) Quantitative RT-PCR analysis of Ccnd3, Rag1, Rag2, and Igκ transcripts comparing results for large pre-B cells and small pre-B cells from wild-type or Bach2-deficient mice. One experiment was performed using three mice for each genotype. For panels B and D, relative expression was normalized by Β2M, and the means and SEM were from four (B) or three (D) mouse samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To investigate the requirement of Bach2 in cell cycle arrest of small pre-B cells, we performed cell cycle analysis in wild-type and Bach2-deficient B cells during B cell development. Compared with wild-type cells, Bach2-deficient small pre-B cells failed to arrest at G1 phase, and more of the mutant cells than wild-type cells were in the S phase (Fig. 5C). Bach2-deficient pro-B cells also showed an increase in the ratio of cells in S phase (Fig. 5C). Interestingly, a larger fraction of Bach2-deficient immature and mature B cells entered S phase than wild-type cells. These results suggest that Bach2 is an important regulator of cell cycle arrest in B cells.
The expressions of genes related to cell cycle and recombination were compared between wild-type and Bach2-deficient pre-B cells. In wild-type cells, the expression of Ccnd3 was lower in small pre-B cells than large pre-B cells, which was again consistent with the cell cycle arrest of the small pre-B cells. Ccnd3 mRNA was unexpectedly lower in Bach2-deficient large pre-B cells than in wild-type cells, and unlike what was seen in wild-type cells, it was not further reduced in small pre-B cells (Fig. 5D). These alterations in Bach2-deficient pre-B cells suggest that cell cycle regulators other than Ccnd3 are also deregulated in the absence of Bach2. In contrast, the expression of Rag1 was normally induced in Bach2-deficient small pre-B cells, whereas Igk transcript tended to be higher in Bach2-deficient small pre-B cells than in wild-type cells. These observations together suggest that a proper coordination of cell cycle arrest and immunoglobulin gene rearrangement in pre-B cells was lost in the absence of Bach2.
Bach2 controls G1 arrest in mature B cells.
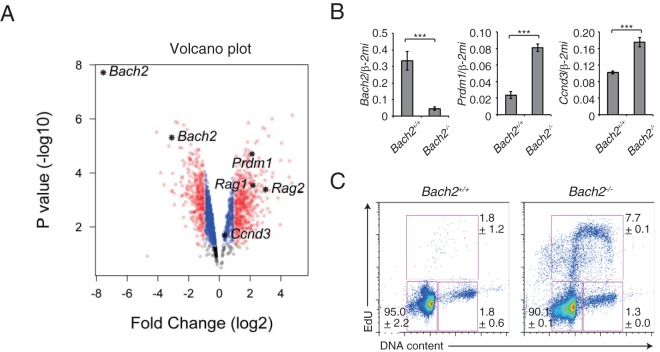
Mature B cells remain dormant until they encounter a cognate antigen. To examine whether this dormancy is affected by Bach2 deficiency, we compared gene expression in splenic naive B cells purified from wild-type and Bach2-deficient splenic B cells and found that the expression of Prdm1 was derepressed in Bach2-deficient B cells in accord with our previous reports (Fig. 6A and B) (15, 16). Surprisingly, we found that the expression of Rag1 and Rag2 was also upregulated in mature B cells, in which these genes are not normally induced (25). Bach2-deficient B cells also showed a slight upregulation of Ccnd3 as well as a striking increase in the ratio of cells in S phase (Fig. 6). Of note, while the number of mature B cell is reduced in Bach2-deficient mice (14), we found a larger number of apoptotic cells in Bach2-deficient mature B cells (see Fig. S7 in the supplemental material). Therefore, these results suggest that Bach2 functions in properly promoting G1 arrest in not only pre-B cells but also mature B cells.
FIG 6.
Abnormal regulation of cell cycle in Bach2-deficient splenic B cells. Comparison analysis of splenic B cells from wild-type or Bach2-deficient mice. (A) Volcano plot for microarray data. (B) Quantitative RT-PCR analysis of Bach2, Ccnd3, and Prdm1 compared in splenic B cells from wild-type or Bach2-deficient mice. Relative expression values were normalized by Β2M, and the means and SEM were from three mice for each genotype in one experiment. ***, P < 0.001. (C) Cell cycle analysis by using flow cytometry. Data are representative of two independent experiments using one or two mice, totalling three mice for each genotype, and shown as means and SEM.
The mTOR-Bach2 axis controls CSR under the BCR signaling cascade.
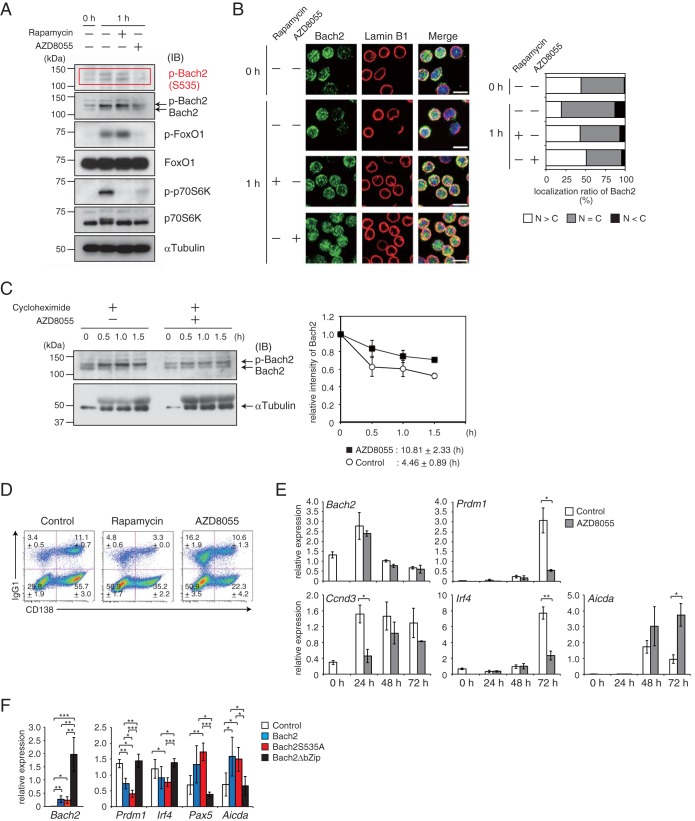
A portion of mature B cells undergo CSR along the way to PC differentiation in response to antigen stimulation, and the frequency of CSR is reduced by the PI3K and mTORC pathways (4, 26). To clarify the importance of Bach2 as a regulatory target of these pathways in the regulation of CSR and PC differentiation, we used splenic B cells isolated from BCR-transgenic mice (27), which can be induced to undergo PC differentiation accompanied with IgG1-type CSR ex vivo, with a higher efficiency than that of wild-type cells (K. Ochiai, submitted for publication). Upon BCR stimulation with NP-Ficoll ex vivo, mTOR was immediately activated, and FoxO1 was phosphorylated in these cells (Fig. 7A). Moreover, the two specific bands of Bach2 were shifted to the slower-migration form, indicating an increased phosphorylation of Bach2. At this time point, the inhibition of mTOR by using rapamycin or AZD8055 blocked the activation of mTOR as judged by the reduction of phosphorylation of p70 S6K. Under these conditions, phosphorylation of FoxO1 was effectively inhibited by AZD8055 but not rapamycin. These observations were similar to the results in pre-B cells, indicating that mTORC2 phosphorylated and inhibited FoxO1 in mature B cells. Two bands of Bach2 were detected with the anti-phosphorylated S535 antibody upon BCR stimulation, and these were reduced only with AZD8055 in a manner similar to that of FoxO1 (Fig. 7A). These observations suggest that both faster and slower bands include Bach2 with phosphorylation at S535. Next, Bach2 protein accumulation was examined by immunohistochemistry. Localization of Bach2 tended to shift from nucleus to cytoplasm by BCR stimulation (Fig. 7B). Treatment with rapamycin or AZD8055 resulted in enhanced nuclear localization of Bach2. Next, we examined the stability of Bach2 protein in these cells. The half-life of endogenous Bach2 protein in the absence of AZD8055 was compared to that in the presence of AZD8055. The faster-migrating Bach2 band disappeared within 1.5 h in the absence of AZD8055, whereas it remained in the presence of AZD8055 (Fig. 7C). Importantly, the half-life of Bach2 was prolonged in the presence of AZD8055 (Fig. 7C). These observations suggest that, similar to IL-7R signaling in pre-B cells, the stability of Bach2 protein is controlled by mTOR downstream of the BCR signal. BCR-mediated activation of mTOR promoted phosphorylation of Bach2 and its cytoplasmic retention and degradation, which are together expected to result in inactivation of Bach2 during PC differentiation.
FIG 7.
Decision of CSR and PC differentiation via the mTORC-Bach2 cascade. Splenic B cells were isolated from B1-8hi mice and stimulated with IL-2, IL-4, IL-5, CD40L, and NP-Ficoll. Cells were cultured for 1 h in the presence of rapamycin or AZD8055 (A and B). (A) Immunoblot analysis for phosphorylated Bach2 at S535 (p-Bach2), total Bach2, phosphorylated FoxO1 (p-FoxO1), FoxO1, p-p70S6K, and p70S6K. α-Tubulin served as an internal control. (B) Immunohistochemistry for Bach2 protein. (Left) Bach2 (green) and lamin B1 (red) distribution in cells. (Right) The subcellular localization of Bach2 was evaluated by classification of 100 cells as described for Fig. 1C. Bar, 10 μm. (C) Half-life of Bach2 protein. Cells were cultured with or without AZD8055 in the presence of cycloheximide under IL-7 Hi conditions. (Left) Immunoblot analysis of Bach2. (Right) Relative intensity of total Bach2 protein. The amounts of protein were normalized with α-tubulin. Data are from one experiment using three mice and are shown as means and SEM. (D) Flow-cytometric analysis of CD138 and IgG1. Data are representative of three mice analyzed in one experiment and are shown as the means and SEM. (E) Quantitative RT-PCR analysis of Bach2, Prdm1, Ccnd3, Irf4, and Aicda for the indicated time points in the presence of AZD. One experiment was performed using three mice. (F) Quantitative RT-PCR analysis of Bach2, Prdm1, Irf4, Pax5, and Aicda in cells transduced with control, WT Bach2, Bach2 S535A, or Bach2ΔbZip expression vectors and sorted based on GFP expression. One experiment was performed using four mice. For panels A and C, α-tubulin served as an internal control. IB, immunoblot. For panels D and F, relative expression was normalized by Β2M, and the means and SEM were from three (E) or four (F) mouse samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The inhibition of only mTORC1 by rapamycin reduced the percentages of both IgG1-positive and CD138-positive cells (Fig. 7D), suggesting that mTORC1 activity is required for both CSR and PC differentiation. In contrast to rapamycin, the inhibition of mTORC1/2 by AZD8055 resulted in a higher frequency of IgG1-positive cells and a reduction of CD138-positive cells (Fig. 7D). To investigate whether Bach2 is involved in the effects of AZD8055, we compared wild-type and Bach2-deficient B1-8hi splenic B cells by staining for IRF4, CD138, and IgG1. While AZD8055 decreased the percentages of IRF4- and CD138-positive plasma cells among wild-type cells, it did not substantially affect plasma cell differentiation of Bach2-deficient cells (see Fig. S8A in the supplemental material). FoxO1 protein was expressed at the same level in wild-type and Bach2-deficient cells (see Fig. S8B in the supplemental material). Therefore, these results indicated that Bach2 was required for the effects of AZD8055.
Upon examining mRNA expression, we found reduced expression of Prdm1 and Irf4 with AZD8055 (Fig. 7E), which was consistent with the inhibition of PC differentiation. While the expression of Aicda was reduced by 72 h after stimulation in control cells, it was sustained at a higher level at 72 h in the presence of AZD8055 (Fig. 7E). Importantly, the expression of Ccnd3 was reduced transiently at 24 h with AZD8055. Since the expression of Bach2 mRNA was not largely affected by the inhibition of mTORC1/2 (Fig. 7E), these effects appeared at least in part due to the nuclear accumulation of Bach2 protein (Fig. 7B). Consistent with this interpretation, transduction of these cells with retroviruses expressing wild-type Bach2 or Bach2 S535A resulted in expected alterations in gene expression. The expression of Prdm1 was reduced by both wild-type Bach2 and Bach2 S535A, with the latter showing a stronger effect (Fig. 7F). Both wild-type Bach2 and Bach2 S535A-transduced cells also reduced the expression of Irf4 and increased that of Pax5 and Aicda. However, there was no significant difference in their effects. These effects were not observed with a transduction of Bach2ΔbZip, which cannot bind to DNA. These results strongly support the idea that the BCR-mTOR cascade controls CSR and PC differentiation by targeting Bach2 protein.
DISCUSSION
In this study, we explored Bach2 regulation under receptors that couple to mTOR signaling in B cells (Fig. 8). By using Irf4−/− Irf8−/− pre-B cells, we found that mTORC1 inhibited Bach2 protein accumulation in the nucleus and promoted its degradation. As our previous report indicated, withdrawal of IL-7 signaling activated FoxO1 (2), which promoted the expression of Bach2 mRNA (Fig. 1D). Inhibition of PI3K was insufficient to promote FoxO1 activity in the presence of IL-7, whereas inhibition of mTORC1 and mTORC2 resulted in FoxO1 activation (Fig. 2C and D). Considering that inhibition of mTORC1 did not promote FoxO1 activation (Fig. 2C and D), IL-7 signaling may associate with mTORC2-FoxO1 regulation in a PI3K-independent manner. Thus, the IL-7R–mTOR cascade regulates Bach2 at both transcriptional and posttranslational steps in B cells. Distinct from these responses in pre-B cells, Bach2 seems to be unresponsive to attenuation of IL-7R signaling in pro-B cells (Fig. 1). This difference between pro-B and pre-B cells may be due to the absence or presence of pre-BCR and other signaling pathways, such as MEK/extracellular signal-regulated kinase (ERK), which are also regulated under pre-BCR. In our recent report, we detected multiple phosphorylation sites on Bach2 protein, which include candidate MEK/ERK consensus sites (observation from reference 17). It is possible that a combination of multiple phosphorylations of Bach2 protein regulates the activity of Bach2. Along B cell differentiation, Bach2 is essential for promoting CSR via inhibiting Prdm1 expression (15). While the occurrence of CSR is controlled under the mTORC2-FoxO1 cascade (26), it is not clear whether FoxO1 directly regulates the expression of mRNA required for CSR. Importantly, we elucidated that the mTOR-Bach2 cascade controlled CSR. Transduction of Bach2 S535A resulted in a lower expression of Prdm1 than that of wild-type Bach2 (Fig. 7F). Importantly, AZD8055 increased CSR in wild-type B cells but not in Bach2-deficient B cells, indicating that Bach2 is a critical effector of mTORC2 for the regulation of CSR. These observations together suggest that FoxO1 may regulate CSR in part by activating the expression of Bach2 in B cells. Thus, the mTORC-Bach2 cascade regulates both pre-B cell differentiation and mature B cell activation.
FIG 8.
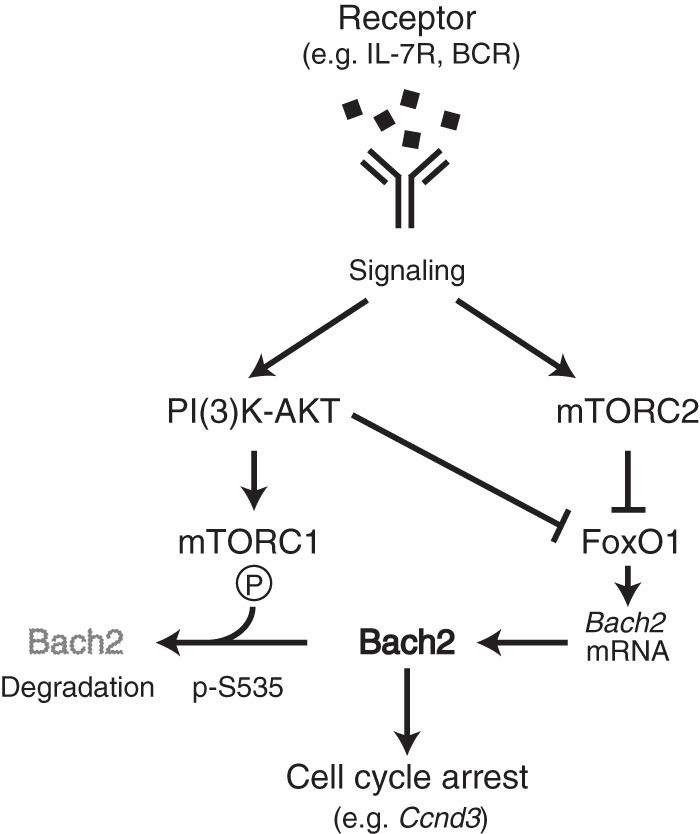
Schematic representation of mTORC-Bach2 cascade in pre-B and mature B cells. Signaling from receptor couples to mTORCs, and Bach2 is regulated under each mTORC1 and mTORC2 at the protein level and transcript level, respectively. mTORC1 mediates Bach2 phosphorylation of serine at 535, which promotes cytoplasmic retention and degradation of Bach2 protein. FoxO1, which is negatively regulated by mTORC2, directly activates Bach2 expression. Therefore, attenuation of receptor signaling reduces mTOR activity, and it enables Bach2 to control the cell cycle by targeting specific genes, such as Ccnd3. Our data suggest that the mTOR-Bach2 cascade functions in pre-B cells with IL-7R and in mature B cells with BCR.
Prdm1 has been well established as a direct target gene of Bach2 in B cells (15–17). In this study, we newly identified Ccnd3 as a direct target gene of Bach2 (Fig. 4), which is known to promote cell cycle transition from G1 to S phase. We showed that Bach2 expression was associated with the reduction of Ccnd3 expression as well as cell cycle arrest in Bach2-RFP pre-B cells (Fig. 5A and B). Furthermore, Bach2-deficient B cells in the bone marrow and spleen showed entry to S phase at stages where normally cells are stalled at G1 phase (Fig. 5C and 6C). Ccnd3 expression was upregulated in Bach2-deficient splenic B cells. These observations indicate that the repression of Ccnd3 by Bach2 contributes to G1 arrest along the course of B cell development. We should note that Ccnd3 expression was lower in Bach2-deficient large pre-B cells than in wild-type cells, and it showed no further reduction in small pre-B cells. Genetic deficiency of Bach2 might have an effect on the basal level of Ccnd3 expression in pre-B cells. Thus, we surmise that Bach2 controls proper cell cycle arrest along B cell development by regulating not only Ccnd3 but also additional regulatory genes of the cell cycle.
During B cell differentiation, DNA rearrangement of immunoglobulin genes is caused by Rag1 and Rag2 in pro-B and pre-B cells. Bach2 regulates the expression of these genes as well. A previous report showed an activation of Rag1/2 gene by Bach2 in BCR-ABL-transformed pre-B cells (18). However, our data showed both a positive and a negative correlation between Bach2 and Rag1/2 expression. As positive correlation, we observed higher expression of Rag1 expression in the Bach2hi pre-B fraction (Fig. 5B). As negative correlation, less nuclear Bach2 localization was accompanied with more Rag1 expression in Rag2−/− pro-B cells (Fig. 1A and C). In addition, Rag1/2 expression was upregulated in Bach2 knockdown Irf4−/− Irf8−/− pre-B cells as well as Bach2-deficient mature B cells (Fig. 1G and 6A). Such contradicting observations might be caused by the modulation of Bach2 by different signaling pathways under receptors. In addition to mTOR, various signaling molecules may tune the interactions of Bach2 with mediators or cofactors, resulting in both activator and repressor functions. Therefore, Bach2 might control Rag1/2 expression in both activation and repression depending on the situations.
We presented the contribution of the mTOR-Bach2 cascades along B cell differentiation. Since recent studies have revealed the importance of Bach2 not only in B cells but also in the differentiation of multipotent progenitors, common lymphoid progenitors, and T-lymphoid cells (10, 13, 28), this cascade may be used in a reiterative manner to control proliferation and differentiation of hematopoietic cells.
MATERIALS AND METHODS
Mice.
Mice used in this study were described previously: Bach2-deficient mice (14), Bach2-RFP mice (10), and BCR transgenic B1-8hi mice (27). All experiments involving mice were approved by the Institutional Animal Care and Use Committee of the Tohoku University Environmental & Safety Committee.
Cells and culture conditions.
Rag2−/− pro-B cells and Irf4−/− Irf8−/− pre-B cells were cultured as described previously (2, 19). Pharmacological inhibitors were from Calbiochem: PI3K inhibitor LY294002, mTORC1 inhibitor rapamycin, and mTORC1/2 inhibitor AZD8055. For experiments using Irf4−/− Irf8−/− pre-B cells, 5 μM LY294002, 20 nM rapamycin, or 500 nM AZD8055 was supplemented in culture medium. For experiments using splenic B cells, cells were isolated as described previously (29). B1-8hi splenic B cells were stimulated with IL-2 (100 U/ml), recombinant mouse IL-4 (5 ng/ml), recombinant mouse IL-5 (1.5 ng/ml), recombinant mouse CD40 ligand (0.2 ng/ml) (all R&D Systems), and NP(40)-Ficoll (0.01 ng/ml; Biosearch Technologies Inc.), and 0.5 nM rapamycin or 10 nM AZD8055 was supplemented in culture medium.
Retroviral vectors and transduction.
Retroviral vectors encoding mouse WT Bach2 (10) or Bach2 S535A (17) with an amino-terminal FLAG tag and green fluorescent protein (GFP) have been reported previously. Retroviral vectors encoding mouse Bach2ΔbZip (in this study) were constructed by subcloning of cDNA segments from pcDNA3 Bach2ΔbZip (30). Plasmids encoding short hairpin RNA oligonucleotides were constructed as described previously (2). The oligonucleotide sequence for short hairpin RNA directed against Bach2 mRNA was 5′-GGGTGCTAAACTTCTACCAAAC-3′. The generation of retroviral stocks was as described previously (2). Cells were suspended in retroviral supernatant supplemented with Polybrene (4 μg/ml) and were centrifuged for 90 min at 2,500 rpm at 32°C. After centrifugation, cells were incubated for 3 h at 37°C and then were washed and cultured under the appropriate conditions. After 48 h, transduced cells were sorted on the basis of GFP expression.
RT-PCR.
For reverse transcription (RT)-PCR, total RNA was prepared by using the RNeasy Plus minikit (Qiagen) or the RNeasy Plus microkit (Qiagen) according to the manufacturer's instructions, and cDNA was synthesized with Superscript III reverse transcriptase (Invitrogen). The SYBR qPCR master mix (Clontech) and Applied Biosystems 7500 Fast real-time PCR system were used for quantitative PCR (for a list of primers, see Table S3 in the supplemental material).
Immunoblot analysis and immunohistochemistry.
Protein extracts were prepared as described previously (2). Each sample was separated by SDS-PAGE and transferred to Immobilon-P membranes (Millipore) and examined by immunoblot analysis as described previously (17). The primary antibodies used were as follows: anti-Bach2 (N1; homemade) and anti-phosphorylated S535 of Bach2 (#17; homemade); anti-FoxO1 (2880), anti-Akt (9272), and anti-p70 S6 kinase (9202) (all from Cell Signaling); antibody to phosphorylated FoxO1 (9461), Akt (4058), and p70 S6 kinase (9205) (all from Cell Signaling); anti-lamin B1 (M-20) (sc-6217), anti-hypoxanthine phosphoribosyltransferase (anti-HPRT) (FL-218) (sc-20975), and anti-α-tubulin (B-7) (sc-5286) (all from Santa Cruz); and anti-FLAG M2-peroxidase (A8592; Sigma). The secondary antibodies used were horseradish peroxidase (HRP)-conjugated anti-rabbit IgG, HRP-conjugated anti-goat IgG, and HRP-conjugated anti-mouse IgG (all from GE Healthcare). Fractionated proteins were isolated by using a subcellular protein fractionation kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. For determination of the half-life of Bach2 protein, bands obtained from immunoblots were scanned and measured by using ImageJ software and normalized by that of HPRT. Immunohistochemistry was performed by using anti-Bach2 (N1) or anti-FoxO1 (9461; Cell Signaling) or anti-lamin B1 (sc-6217; Santa Cruz) as described previously (2).
Flow cytometry.
Cells were washed in staining buffer and phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin (BSA) and 0.05% sodium azide and were blocked with Fc receptor (2.4G2; Tombo Bioscience). Cells were stained with biotin- or fluorescence-coupled antibodies in staining buffer using standard procedures. Anti-mouse mouse antibodies specific for B220 (RA3-6B2), CD43 (S7), IgM (II/41), IgG1 (A85-1), and CD138 (281-2) were from BD Pharmingen. Each population of B cell differentiation stage is gated on the basis of surface marker as follows: pro-B (B220+ CD43hi IgM−), pre-B (B220+ CD43lo IgM−), immature and mature B in bone marrow (B220+ CD43lo IgM+); and large and small pre-B cells were gated with side scatter (SSC) and forward scatter (FSC) for cell size. Mature B cells in spleen were gated with B220+. For intracellular staining, washed cells were then fixed by incubation for 10 min at 25°C with 1% paraformaldehyde and permeabilized in staining buffer containing 0.03% saponin (Fluka). Staining buffer containing 0.3% saponin was used to incubate the cells with normal rabbit IgG (sc-2027; Santa Cruz) or anti-FoxO1 antibody (sc-11350; Santa Cruz) or anti-IRF4 (sc-6069; Santa Cruz) in the presence of 5% donkey serum (Jackson ImmunoResearch). Labeling was detected using a Cy5-coupled donkey anti-goat antibody (Jackson ImmunoResearch). For cell cycle analysis, the Click-iT EdU Alexa Fluor 488 flow cytometry assay kit (C10425; Thermo Fisher) was used according to the manufacturer's instructions. Data were collected with the FACSCanto II system and analyzed with FlowJo software (TreeStar).
Microarray analysis.
Total RNAs were prepared and labeled with Cy3 by using the low input quick amp labeling kit (Agilent). A SurePrint G3 mouse GE microarray slide was used according to the manufacturer's instructions (Agilent). The data were detected on an Agilent scanner. Data were analyzed with the GeneSpring software package (Agilent).
ChIP assay.
ChIP was performed as previously described (2). Chromatin from Irf4−/− Irf8−/− pre-B cells was isolated and then sonicated to obtain DNA fragments ranging in size from 100 to 500 bp. Chromatin fragments were immunoprecipitated with normal rabbit IgG (sc-2027; Santa Cruz), anti-FoxO1 (sc-11350; Santa Cruz), anti-Bach2 antiserum (N1; homemade), or anti-MafK (sc-477; Santa Cruz). After reversal of formaldehyde cross-links, specific DNA sequences were assessed by quantitative real-time PCR. Primers used in ChIP PCR are shown in Table S3.
Statistical analysis.
Statistical analysis was performed with the Welch two-sample t test using the open-source statistical programming environment R.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Igarashi laboratory for helpful discussions and T. Kurosaki (Osaka University) and M. Nussenzweig (Rockfeller University) for providing BCR transgenic B1-8hi mice.
This study was supported by grants-in-aid from the Ministry of Education, Science, Sport, and Culture of Japan (KAKENHI 16K15227, 15H02506, 24390066, 16K19026, 16H01295) and AMED-CREST. This project was supported by Astellas Foundation for Research on Metabolic Disorders (K.O.). This work was supported in part by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from AMED from Japan Agency for Medical Research and development, AMED (Y.K.).
K.O. and K.I. designed and T.T. performed the majority of the experiments with assistance from K.O. K.O. performed the ChIP assay, the analysis of Bach2-RFP mice by using flow cytometry, the microarray of the comparison between wild-type versus Bach2-deficient splenic B cells, the immunoblot analysis of retrovirus transduction using B1-8hi splenic B cells, and the experiments using Bach2-deficient splenic B cells with B1-8hi background. A.M. supported the screening of Bach2 binding sites in Ccnd3 and Ccne locus. Y.K. assisted A.M. in designing and generating the specific antibodies against Bach2 S535 phosphorylation. N.S. assisted A.M. in experiments. M.M. supported with data analysis the microarray that K.O. performed. T.K. contributed to analysis and evaluation of data. T.T., K.O., and K.I. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00418-17.
REFERENCES
- 1.Clark MR, Mandal M, Ochiai K, Singh H. 2014. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol 14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. 2012. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol 13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rickert RC. 2013. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol 13:578–591. doi: 10.1038/nri3487. [DOI] [PubMed] [Google Scholar]
- 4.Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, Calame K, Rickert RC. 2006. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity 25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Aoki M, Jiang H, Vogt PK. 2004. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc Natl Acad Sci U S A 101:13613–13617. doi: 10.1073/pnas.0405454101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhuang G, Yu K, Jiang Z, Chung A, Yao J, Ha C, Toy K, Soriano R, Haley B, Blackwood E, Sampath D, Bais C, Lill JR, Ferrara N. 2013. Phosphoproteomic analysis implicates the mTORC2-FoxO1 axis in VEGF signaling and feedback activation of receptor tyrosine kinases. Sci Signal 6:ra25. doi: 10.1126/scisignal.2003572. [DOI] [PubMed] [Google Scholar]
- 7.Asada S, Daitoku H, Matsuzaki H, Saito T, Sudo T, Mukai H, Iwashita S, Kako K, Kishi T, Kasuya Y, Fukamizu A. 2007. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell Signal 19:519–527. doi: 10.1016/j.cellsig.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Amin RH, Schlissel MS. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol 9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson K, Chaumeil J, Micsinai M, Wang JM, Ramsey LB, Baracho GV, Rickert RC, Strino F, Kluger Y, Farrar MA, Skok JA. 2012. IL-7 functionally segregates the pro-B cell stage by regulating transcription of recombination mediators across cell cycle. J Immunol 188:6084–6092. doi: 10.4049/jimmunol.1200368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh-Nakadai A, Hikota R, Muto A, Kometani K, Watanabe-Matsui M, Sato Y, Kobayashi M, Nakamura A, Miura Y, Yano Y, Tashiro S, Sun J, Ikawa T, Ochiai K, Kurosaki T, Igarashi K. 2014. The transcription repressors Bach2 and Bach1 promote B cell development by repressing the myeloid program. Nat Immunol 15:1171–1180. doi: 10.1038/ni.3024. [DOI] [PubMed] [Google Scholar]
- 11.Igarashi K, Itoh-Nakadai A. 2016. Orchestration of B lymphoid cells and their inner myeloid by Bach. Curr Opin Immunol 39:136–142. doi: 10.1016/j.coi.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Kuwahara M, Suzuki J, Tofukuji S, Yamada T, Kanoh M, Matsumoto A, Maruyama S, Kometani K, Kurosaki T, Ohara O, Nakayama T, Yamashita M. 2014. The Menin-Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis. Nat Commun 5:3555. doi: 10.1038/ncomms4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, Ji Y, Sukumar M, Eil RL, Yu Z, Spolski R, Palmer DC, Pan JH, Patel SJ, Macallan DC, Fabozzi G, Shih HY, Kanno Y, Muto A, Zhu J, Gattinoni L, O'Shea JJ, Okkenhaug K, Igarashi K, Leonard WJ, Restifo NP. 2016. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 17:851–860. doi: 10.1038/ni.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. 2004. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 15.Muto A, Ochiai K, Kimura Y, Itoh-Nakadai A, Calame KL, Ikebe D, Tashiro S, Igarashi K. 2010. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J 29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, Igarashi K. 2006. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem 281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 17.Ando R, Shima H, Tamahara T, Sato Y, Watanabe-Matsui M, Kato H, Sax N, Motohashi H, Taguchi K, Yamamoto M, Nio M, Maeda T, Ochiai K, Muto A, Igarashi K. 2016. The transcription factor Bach2 is phosphorylated at multiple sites in murine B cells but a single site prevents its nuclear localization. J Biol Chem 291:1826–1840. doi: 10.1074/jbc.M115.661702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, Ng C, Titz B, Hurtz C, Sadiyah MF, Nowak D, Thoennissen GB, Rand V, Graeber TG, Koeffler HP, Carroll WL, Willman CL, Hall AG, Igarashi K, Melnick A, Muschen M. 2013. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med 19:1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertolino E, Reddy K, Medina KL, Parganas E, Ihle J, Singh H. 2005. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol 6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 20.Lu R, Medina KL, Lancki DW, Singh H. 2003. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev 17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity 28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida C, Yoshida F, Sears DE, Hart SM, Ikebe D, Muto A, Basu S, Igarashi K, Melo JV. 2007. Bcr-Abl signaling through the PI-3/S6 kinase pathway inhibits nuclear translocation of the transcription factor Bach2, which represses the antiapoptotic factor heme oxygenase-1. Blood 109:1211–1219. doi: 10.1182/blood-2005-12-040972. [DOI] [PubMed] [Google Scholar]
- 23.Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H, Clark MR. 2009. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol 10:1110–1117. doi: 10.1038/ni.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper AB, Sawai CM, Sicinska E, Powers SE, Sicinski P, Clark MR, Aifantis I. 2006. A unique function for cyclin D3 in early B cell development. Nat Immunol 7:489–497. doi: 10.1038/ni1324. [DOI] [PubMed] [Google Scholar]
- 25.Yu W, Nagaoka H, Jankovic M, Misulovin Z, Suh H, Rolink A, Melchers F, Meffre E, Nussenzweig MC. 1999. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature 400:682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 26.Limon JJ, So L, Jellbauer S, Chiu H, Corado J, Sykes SM, Raffatellu M, Fruman DA. 2014. mTOR kinase inhibitors promote antibody class switching via mTORC2 inhibition. Proc Natl Acad Sci U S A 111:E5076–E5085. doi: 10.1073/pnas.1407104111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih TA, Meffre E, Roederer M, Nussenzweig MC. 2002. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol 3:570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- 28.Itoh-Nakadai A, Matsumoto M, Kato H, Sasaki J, Uehara Y, Sato Y, Ebina-Shibuya R, Morooka M, Funayama R, Nakayama K, Ochiai K, Muto A, Igarashi K. 2017. A Bach2-Cebp gene regulatory network for the commitment of multipotent hematopoietic progenitors. Cell Rep 18:2401–2414. doi: 10.1016/j.celrep.2017.02.029. [DOI] [PubMed] [Google Scholar]
- 29.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong AS, Klein U, Dinner AR, Singh H, Sciammas R. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38:918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochiai K, Muto A, Tanaka H, Takahashi S, Igarashi K. 2008. Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int Immunol 20:453–460. doi: 10.1093/intimm/dxn005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.