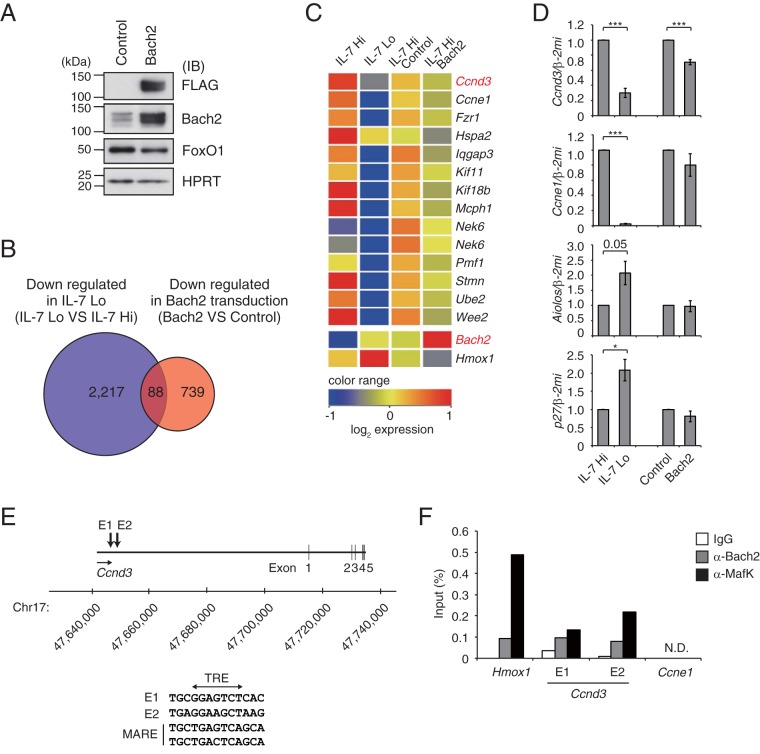
FIG 4.
Ccnd3 gene as a putative target of Bach2. (A) Immunoblot analysis of indicated proteins in Irf4−/− Irf8−/− pre-B cells transduced with control or Bach2 expression vector and then sorted based on GFP expression. HPRT served as an internal control. (B) Venn diagram of microarray analysis in Irf4−/− Irf8−/− pre-B cells comparing downregulated genes with IL-7 attenuation (blue) and Bach2 transduction with IL-7 Hi (red). Among 2,305 genes downregulated with IL-7 attenuation (21), 88 genes overlapped genes that were downregulated by Bach2 transduction. (C) Heatmap of cell cycle-related gene expression in a comparison of IL-7 Hi and IL-7 Lo conditions and of transduction of control vector versus Bach2. Transcripts of Hmox1 were shown to be one of the direct targets of Bach2. (D) Quantitative RT-PCR analysis of Ccnd3, Ccne1, Aiolos, and p27 in Irf4−/− Irf8−/− pre-B cells as described for panel C. Relative expression was normalized by Β2M, and the means and SEM were from three independent experiments. *, P < 0.05; ***, P < 0.001. (E) MARE-like sequences E1 and E2 at Ccnd3 locus. Bach2 binding sequences detected in these regions were shown along with MARE consensus. E, enhancer. (F) Binding of Bach2 and MafK to regulatory elements in the Hmox1, Ccnd3 E1 and E2, and Ccne1 loci in cells cultured with 0.1 ng/ml of IL-7 for 48 h, assessed by ChIP with control IgG, anti-Bach2, or anti-MafK. The data are the averages from three independent experiments. N.D., not detected.