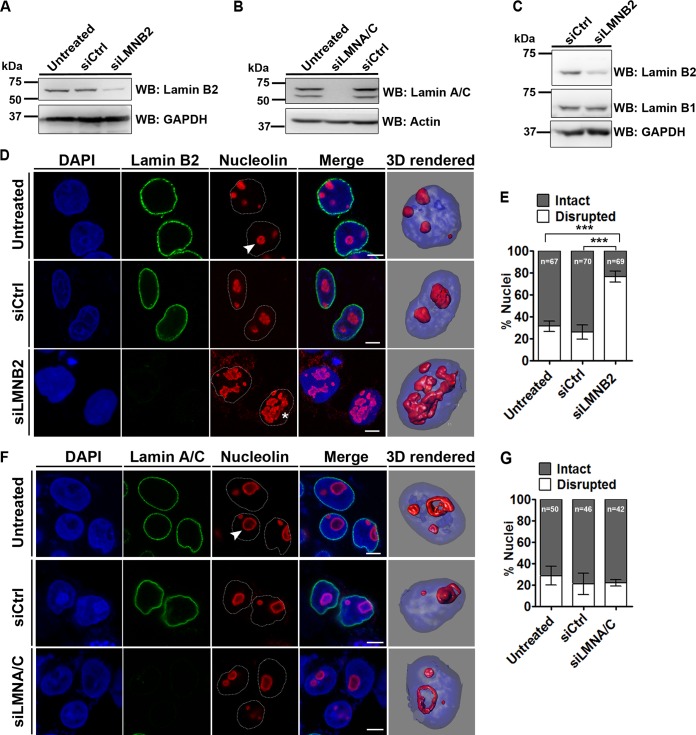
FIG 1.
Lamin B2 depletion disrupts nucleolar morphology. (A and B) Western blots (WB) showing siRNA-mediated depletion of lamin B2 (A) and lamin A/C (B). (C) Lamin B1 levels are unaltered upon lamin B2 knockdown. Loading controls were GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and actin. (D) Coimmunostaining of lamin B2 and nucleolin in DLD-1 cells. DAPI counterstains the nucleus. Untreated cells, lamin B2 knockdown cells (siLMNB2), or cells treated with nontargeting siRNA (siCtrl) were used. Representative confocal images of control cells show discrete and intact nucleoli (arrowhead), and siLMNB2 shows disrupted nucleolar morphology (asterisk) (also shown in 3D reconstructions). Scale bars, ∼5 μm. (E) Quantification of nucleolar morphologies shows a significant increase in disrupted nucleoli upon lamin B2 knockdown (***, P < 0.001 by Fisher's exact test of proportions) (number of independent biological replicates [N] = 3; n, number of nuclei). Error bars indicate standard errors of means (SEM). (F) Coimmunostaining of lamin A/C and nucleolin. No change in nucleolar morphology was observed upon siLMNA/C (arrowhead) or siCtrl treatment (also shown in 3D reconstructions). Scale bars, ∼5 μm. (G) Quantification of nucleolar morphologies upon lamin A/C knockdown (P > 0.05 by Fisher's exact test of proportions) (N = 2; n, number of nuclei). Error bars indicate standard deviations (SD).