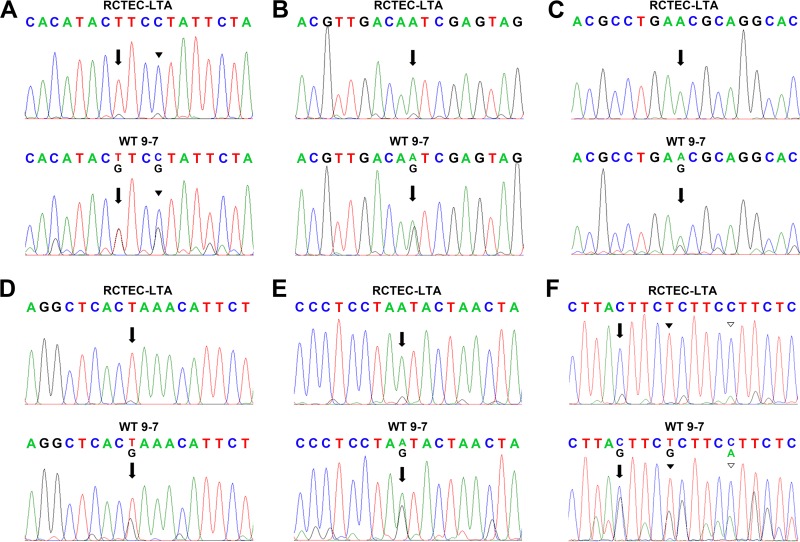
FIG 7.
Comparison of mtDNA sequences in normal tubular cells (RCTEC-LTA) and cyst-derived cells with a heterozygous PKD1 mutation (WT 9-7). Representative examples of mtDNA sequences, with accumulated mtDNA point mutations and heteroplasmy in cyst-derived cells, are shown. The influence of missense mutations on protein function was analyzed using polyPhen-2. (A) The region encoding NADH dehydrogenase subunit 4. The arrows point to mtDNA position 11204, and the arrowheads point to position 11207. A T-to-G replacement at position 11204 translates to a phenylalanine-to-valine mutation predicted to be benign (polyPhen-2 score, 0.035). A C-to-G replacement at position 11207 translates to a leucine-to-valine mutation predicted to be damaging (polyPhen-2 score, 0.994). (B) The region encoding cytochrome c oxidase subunit 2. The arrows show mtDNA position 8004. Replacement of A by G at position 8004 alters the amino acid asparagine to serine, and this mutation is predicted to be probably damaging, with a polyPhen-2 score of 0.973. (C) The region encoding NADH dehydrogenase subunit 4. The arrows show mtDNA position 11190. Replacement of A by G at position 11190 alters the amino acid asparagine to serine, which is predicted to be probably damaging, with a polyPhen-2 score of 0.994. (D) The region encoding NADH dehydrogenase subunit 4. The arrows show mtDNA position 11280. Replacement of T by G at position 11280 alters the amino acid leucine to arginine, which is predicted to be probably damaging, with a polyPhen-2 score of 0.999. (E) The region encoding NADH dehydrogenase subunit 4. The arrows show mtDNA position 11958. Replacement of A by G at position 11958 alters the amino acid methionine to serine, which is predicted to be benign, with a polyPhen-2 score of 0.121. (F) The region encoding cytochrome b. The arrows show mtDNA position 15443. Replacement of C by G at position 15443 alters the amino acid leucine to valine, which is predicted to be benign, with a polyPhen-2 score of 0.010. The black arrowhead shows mtDNA position 15447. Replacement of T by G at position 15447 alters the amino acid leucine to proline, which is predicted to be benign, with a polyPhen-2 score of 1.000. The white arrowhead shows mtDNA position 15452. Replacement of G by A at position 15452 alters the amino acid leucine to phenylalanine, which is predicted to be benign, with a polyPhen-2 score of 1.000.