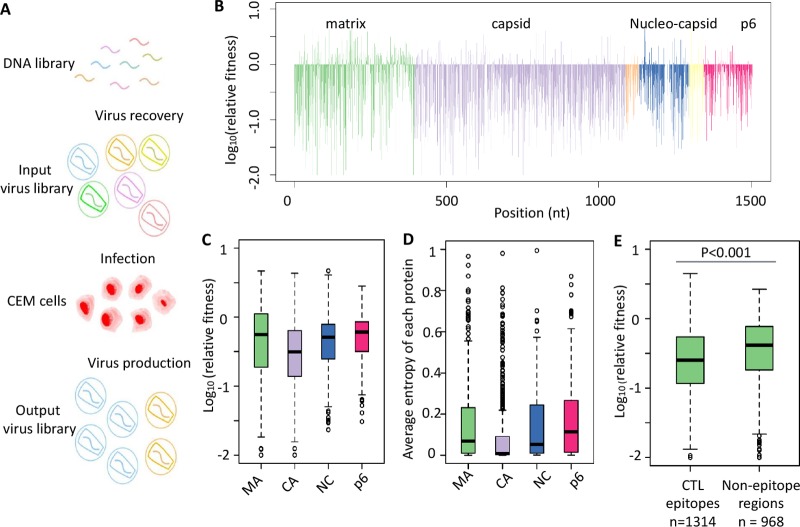
FIG 1 .
Quantitative high-throughput fitness profiling of HIV-I Gag polyprotein. (A) Experimental design of high-throughput fitness profiling of the HIV-1 Gag region. (B) Relative fitness score of each point mutation in Gag. Each Gag protein is labeled by a different color. MAb, matrix; CA, capsid; SP1, spacer peptide 1; NC, nucleocapsid; SP2, spacer peptide 2; p6, p6 protein; nt, nucleotide. (C) Average relative fitness scores of missense mutations in each Gag protein are shown using a box plot. (D) The average entropy of each Gag protein was calculated based on naturally occurred variants in the HIV sequence database at Los Alamos National Laboratory. (E) Relative fitness scores of mutations within or outside CTL epitope regions were compared. CTL epitopes were defined according to the 2013 update of best-characterized epitopes from the Los Alamos Database. A total of 1,314 mutations within CTL epitopes and a total of 968 mutations outside CTL epitopes were calculated.