FIG 2 .
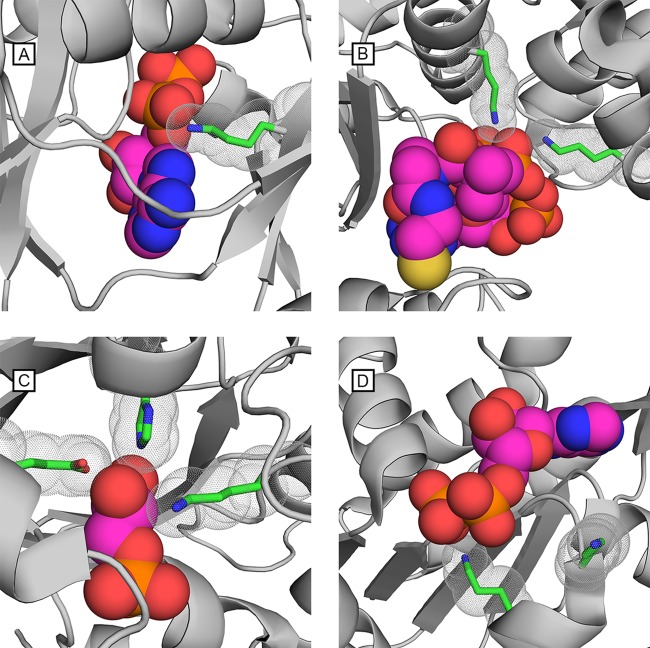
Substrate-binding lysine residues in metabolic enzymes are evolutionarily conserved and acetylated in different organisms. In many glycolytic and TCA cycle catalytic binding sites, lysine is present and universally conserved. The binding pockets of four enzymes are shown with their native substrate or cofactor. Catalytically required lysine residues are shown and outlined with spheres to show their proximity to the substrate or cofactor. (A) Succinyl-CoA synthetase from E. coli, PDB 1CQI, shown with the ADP cofactor and lysine 46. (B) Phosphotransacetylase from Methanosarcina thermophila, PDB 2AF4, shown with coenzyme A and two lysine residues 257 and 283. (C) Triosephosphate isomerase from Staphylococcus aureus, PDB 3UWU, shown with glycerol-3-phosphate and lysine 12. Catalytically essential histidine and glutamic acid residues are also shown. (D) Phosphoglycerate kinase from Francisella tularensis, PDB 4FEY, shown with the ADP cofactor and lysine residues 193 and 197.