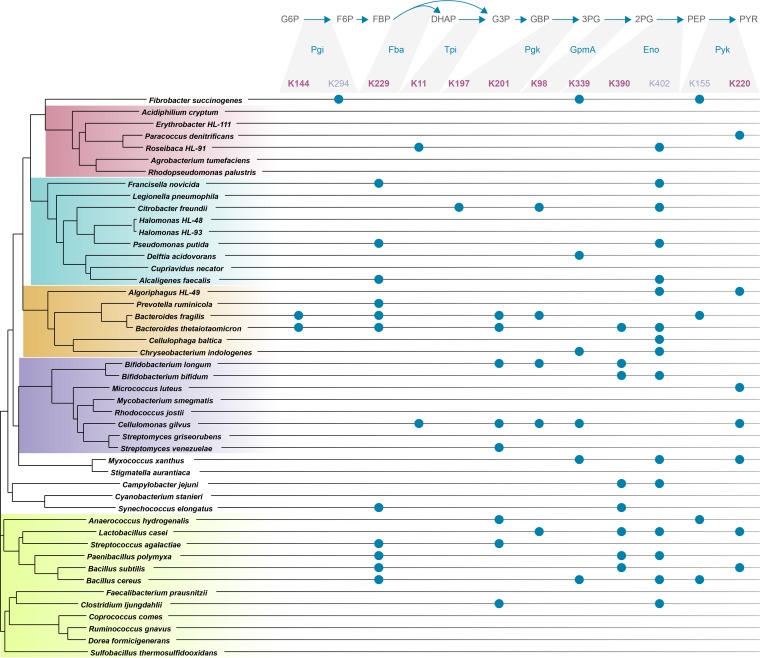
FIG 4 .
Acetylation of substrate/cofactor-binding lysine residues across taxa. The phyloproteomic data identify acetylated lysines from 48 organisms. Enzymes from glycolysis are listed along with universally conserved lysine residues, numbered according to the numbering system for Bacillus subtilis (except for GpmA from L. casei gene S6CBB3). Catalytically essential sites as described in the text are shown in boldface type. Nonboldfaced sites are listed if they are universally conserved across bacteria but are not known to be involved in substrate/cofactor binding. Observed acetylations are indicated with solid blue circles. The phylogenetic tree of organisms is based on RplB sequence alignment with the major taxonomic groups colored: Alphaproteobacteria, Gamma- or Betaproteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes. Protein name abbreviations: Pgi, phosphoglucose isomerase; Fba, fructose-bisphosphate aldolase; Tpi, triose-phosphate isomerase; Pgk, phosphoglycerate kinase; GmpA, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase; Eno, enolase; Pyk, pyruvate kinase.