Abstract
Objective
The aim of the present study is to optimize the PCR conditions required to amplify the promoter sequence of an amino acid transporter having an AT-rich base composition with a high number of tandem repeats.
Result
Results show that successful amplification can be achieved by performing a 2-step PCR at a lower extension temperature of 65 °C for an increased extension period of 1.5 min/kb, with MgCl2 concentration ranging from 2.5 to 3.0 mM. The results also suggest that the DNA concentration of about 25–30 ng/µl was essential to achieve this amplification.
Keywords: AT-rich, PCR, Optimization, Tandem repeats, Extension
Introduction
PCR is one of the indispensable techniques in molecular biology for in vitro amplification of a specific segment of DNA [1]. It is highly reliable because of its sensitivity, accuracy, and speed [2, 3]. However, under specific requirements such as to amplify templates, which are AT- or GC-rich, or have a high number of tandem repeats the PCR conditions need to be optimized. Plant promoter regions are generally difficult to amplify by PCR as they are highly AT-rich and sometimes contain tandem repetitive DNA sequences [4, 5]. Tandem repeats represent two or more copies of short segments of DNA occurring repeatedly from head-to-tail within the coding and regulatory regions [6]. The problem with these templates is that they need lower annealing and extension temperatures which can result in the amplification of undesired products [7, 8]. The aim of this work is to amplify a promoter sequence (1781 bp) of an amino acid transporter (AT2G40420) from Arabidopsis thaliana, which is highly AT-rich and has a high number of tandem repeats.
The in silico analysis of the promoter sequence reveals that it possesses many important cis-acting regulatory elements such as light-responsive, auxin-responsive, salicylic acid-responsive, and abscisic acid-responsive elements along with 16 copies of an ACGT motif [9]. Studies suggest that the cis-elements with ACGT core sequence responds to light, anaerobiosis, phytohormones like abscisic acid, jasmonic acid, salicylic acid and auxin [10, 11]. Furthermore, Zou et al. [12] conducted a study where they concluded that around 19.6% of the total pCREs (putative cis-regulatory elements) identified in the promoter regions of abiotic stress responsive genes have ACGT as a core sequence. Therefore, tapping this promoter sequence for its response to abiotic stress conditions can potentially bring forth important characteristics that can further find wide application for generation of transgenic plants with high stress tolerance. As, a suitable promoter is needed to achieve desired expression levels of a transgene [13]. In the study, the promoter sequence (AT2G40420, 1781 bp) was amplified from Arabidopsis thaliana genome. However, the sequence is 65.2% AT-rich and has 15.5 copies of 28 base long tandem repeat [14], which makes it difficult to amplify by PCR (Fig. 1). These tandem repeat sequences have a binding site for bZIP (basic leucine zipper) transcription factors (TFs). Reports suggest that tandem repeats possessing binding sites for transcription factors in the promoter regions can affect the transcriptional rate of a gene [15]. To check the effect of all these TF binding sites localized in tandem repeats on the downstream gene expression, isolation of the promoter sequence with all the copies of tandem repeats was highly desirable.
Fig. 1.
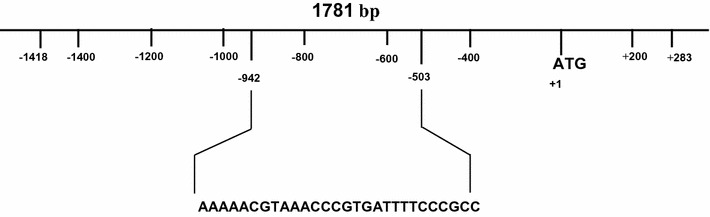
Amino acid transporter (AT2G40420) promoter region (1781 bp). The sequence and position of 28 bp long tandem repeat, occurring 15.5 times in the promoter region from − 503 to − 942 and the translation start site ATG, are depicted in the figure
Main text
Methods
Plant material and growth conditions
Arabidopsis thaliana, ecotype Columbia (Col-0) was used in this study. Arabidopsis seeds were procured from LEHLE SEEDS Company (Catalog number: WT-02), Texas, USA. Seeds were vernalized at 4 °C in the dark for 3 days before sowing in 5.08 cm plastic pots filled with soilrite. Thereafter, the pots were transferred into a growth chamber (Daihan Labtech, LGC-5101, India) maintaining a 16-h light/8-h dark photoperiod, 22 °C temperature, 75% relative humidity. After 3 weeks, rosette leaves were harvested to extract the genomic DNA.
Genomic DNA isolation
Genomic DNA was isolated from the leaves of Arabidopsis thaliana (ecotype Col-0) using the DNeasy Plant Mini Kit (Qiagen, Cat No./ID: 69104) according to the manufacturer’s recommendations. The DNA integrity was confirmed by running 0.8% agarose gel electrophoresis at 80 V for 30 min.
Primer design
Primers were designed to amplify a 1781 bp promoter sequence of the amino acid transporter (AT2G40420) (Table 1) using the Primer3 program [16]. Their specificity was ensured by performing primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) with the Arabidopsis genome. Further, the OligoAnalyzer tool supported by Integrated DNA Technologies (https://www.idtdna.com/SciTools/SciTools.aspx.) was used to check for the presence of any secondary structure or primer–dimer formation.
Table 1.
Forward and reverse primer specifications for AT2G40420 promoter sequence
| Primer | Primer sequence (5′→3′) | Tm °C | GC% | Product size |
|---|---|---|---|---|
| AT2G40420F | CCTACTAGTTCGTGATACTG | 52.05 | 45.00 | 1781 bp |
| AT2G40420R | CGAACGATTCCTTCATCACG | 57.02 | 50.00 |
PCR conditions
Each 20 µl PCR contained 2 μl of genomic DNA ( ~ 50 ng), 4 µl of 5X Phusion HF buffer, 0.4 µl of 10 mM dNTPs, 0.8 µl of each 10 µM forward and reverse primer, 0.2 µl of Phusion DNA polymerase (2U/µl), and varying concentrations of MgCl2 ranging from 1.5 to 3.5 mM. All the reagents were procured from Thermo Fisher SCIENTIFIC (Catalog number: F530S, Waltham, MA, USA) and MB grade nuclease-free water from Himedia (Catalog number: ML024). A 2-step PCR was carried out using the Applied Biosystems® Veriti® 96-Well Thermal Cycler (Catalog number: 4375786, Foster City, CA, USA) with conditions as follows: Initial denaturation at 98 °C for 1.5 min; followed by 35 cycles of denaturation at 98 °C for 30 s, extension at 60/65/68/72 °C for 3 min and final extension at 60/65/68/72 °C for 7 min. PCR for each extension temperature with varying MgCl2 concentrations were performed separately and in triplicates. PCR products were checked by electrophoresis in 1% (w/v) agarose gel, at 80 V for 30 min.
Amplicon sequence analyses
The QIAquick Gel Extraction Kit (Qiagen, Catalog number: 28704) was used to purify the PCR products. The purified PCR product along with the primers used for amplification, was then directed for sequencing to verify the specificity of the amplified product. The amplicon specificity was confirmed by analysing the obtained sequencing results with the reference sequence deposited in the TAIR database (https://www.arabidopsis.org) of the amino acid transporter promoter region [17].
Results and discussion
Concentration of magnesium ions
The magnesium ion concentration greatly influences the PCR as DNA polymerase requires Mg2+ ions for its proper functioning [18, 19]. Therefore, to achieve maximal PCR yield the MgCl2 concentration needs to be optimized. As, a high Mg2+ ion concentration can hinder the reaction by preventing proper melting of template DNA and can also promote non-specific binding of primers. Even a low Mg2+ ion concentration can adversely affect the product yield. With this aim, varying concentrations of MgCl2 such as 1.5, 2.0, 2.5, 3.0, 3.5 mM were tried. The desired amplicon yield was obtained at a 3.0 mM MgCl2 concentration (Fig. 2).
Fig. 2.
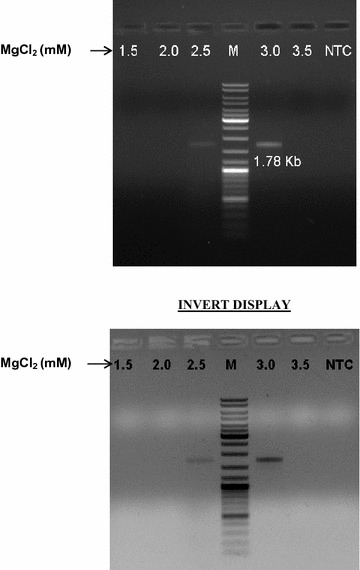
Effects of MgCl2 concentration on PCR amplification at an extension temperature of 65 °C. Lane M: 10 kb DNA ladder; lane 1: 1.5 mM MgCl2; lane 2: 2 mM MgCl2; lane 3: 2.5 mM MgCl2; lane 5: 3 mM MgCl2; lane 6: 3.5 mM MgCl2; lane 7: no-template negative control
Extension temperature
For successful amplification, the extension time and temperature need to be carefully optimized. Xin-Zhuan Su et al. [20] reported that to amplify an AT-rich DNA, reduced extension temperatures are needed. In the present study, a two-step PCR (denaturation and amplification) was performed at four different extension temperatures 60, 65, 68 and 72 °C with increased extension time from the usual 1 to 1.5 min/kb. Successful amplification was achieved at an extension temperature of 65 °C with 2.5 mM MgCl2 yielding a faint band while an intense band was observed with 3 mM MgCl2 concentration (Fig. 2). No results were obtained at other extension temperatures (60, 68, and 72 °C) at any of the five MgCl2 concentrations tested (data not shown).
Conclusions
The promoter region of the amino acid transporter was difficult to amplify by PCR owing to its high AT content and a high number of tandem repeats. Successful amplification can be achieved, after optimization of MgCl2 concentration and extension temperature with the DNA template of desired concentration.
Limitations
The DNA template should be pure, homogeneous and concentration should be around 50–60 ng for setting up a 20 µl PCR reaction.
Efficient for AT-rich DNA templates.
Author’s contributions
PD designed, performed the experiments and wrote the manuscript. RM and SM gave critical inputs on designing of experiments, supervised the study and corrected the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to the Birla Institute of Technology and Science, Pilani, Rajasthan, India, for providing the infrastructural and logistic support needed for this work.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
PD is thankful to the Birla Institute of Technology & Science, Pilani for the fellowship. RM and SM are thankful to Department of Science and technology for financial support. This work was supported by SERB project EMR/2016/002470 sanctioned by the government of India to SM and RM.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- dNTP
deoxyribonucleotide triphosphate
- PCR
polymerase chain reaction
- TRs
tandem repeats
- TFs
transcription factors
- pCREs
putative cis-regulatory elements
Contributor Information
Pinky Dhatterwal, Email: pdhatterwal.11@gmail.com.
Sandhya Mehrotra, Email: sandhya@pilani.bits-pilani.ac.in.
Rajesh Mehrotra, Phone: +91-9983734134, Phone: +91-1596-245669, Email: rmehrotra@pilani.bits-pilani.ac.in.
References
- 1.Garibyan L, Avashia N. Polymerase chain reaction. J Invest Dermatol. 2013;133:3. doi: 10.1038/jid.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman WB, Tsongalis GJ. The polymerase chain reaction. In: Coleman WB, Tsongalis GJ, editors. Molecular diagnostics for the clinical laboratorian. Totowa: Humana Press Inc; 2006. pp. 47–55. [Google Scholar]
- 3.Obradovic J, Jurisic V, Tosic N, Mrdjanovic J, Perin B, Pavlovic S, et al. Optimization of PCR conditions for amplification of GC-rich EGFR promoter sequence. J Clin Lab Anal. 2013;27:487–493. doi: 10.1002/jcla.21632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahdev S, Saini S, Tiwari P, Saxena S, Saini KS. Amplification of GC-rich genes by following a combination strategy of primer design, enhancers and modified PCR cycle conditions. Mol Cell Probes. 2007;21:303–307. doi: 10.1016/j.mcp.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Gemayel R, Cho J, Boeynaems S, Verstrepen KJ. Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes. 2012;3:461–480. doi: 10.3390/genes3030461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quilez J, Guilmatre A, Garg P, Highnam G, Gymrek M, Erlich Y, et al. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res. 2016;44:3750–3762. doi: 10.1093/nar/gkw219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hommelsheim CM, Frantzeskakis L, Huang M, Ülker B. PCR amplification of repetitive DNA: a limitation to genome editing technologies and many other applications. Sci Rep. 2014;4:5052. doi: 10.1038/srep05052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy S, Oswald N. PCR troubleshooting and optimization: the essential guide. Norfolk: Caister Academic Press; 2011. [Google Scholar]
- 9.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehrotra R, Yadav A, Bhalothia P, Karan R, Mehrotra S. Evidence for directed evolution of larger size motif in Arabidopsis thaliana genome. Sci World J. 2012; 1–5. 10.1100/2012/983528. [DOI] [PMC free article] [PubMed]
- 11.Mehrotra R, Sethi S, Zutshi I, Bhalothia P, Mehrotra S. Patterns and evolution of ACGT repeat cis-element landscape across four plant genomes. BMC Genom. 2013;14:203. doi: 10.1186/1471-2164-14-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou C, Sun K, Mackaluso JD, Seddon AE, Jin R, Thomashow MF, Shiu SH. Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2011;108:14992–14997. doi: 10.1073/pnas.1103202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potenza C, Aleman L, Sengupta-Gopalan C. Invited review: targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. InVitro Cell Dev Biol Plant. 2004;40:1–22. doi: 10.1079/IVP2003477. [DOI] [Google Scholar]
- 14.Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, et al. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016;44:D1154–D1160. doi: 10.1093/nar/gkv1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, et al. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekman S. PCR optimization and troubleshooting, with special reference to the amplification of ribosomal DNA in lichenized fungi. Lichenologist. 1999;31:517–531. doi: 10.1017/S0024282999000675. [DOI] [Google Scholar]
- 19.Cao Y, Zheng Y, Fang B. Optimization of polymerase chain reaction-amplified conditions using the uniform design method. J Chem Technol Biotechnol. 2004;79:910–913. doi: 10.1002/jctb.1078. [DOI] [Google Scholar]
- 20.Su XZ, Wu Y, Sifri CD, Wellems TE. Reduced extension temperatures required for PCR amplification of extremely A+T-rich DNA. Nucleic Acids Res. 1996;24:1574–1575. doi: 10.1093/nar/24.8.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


