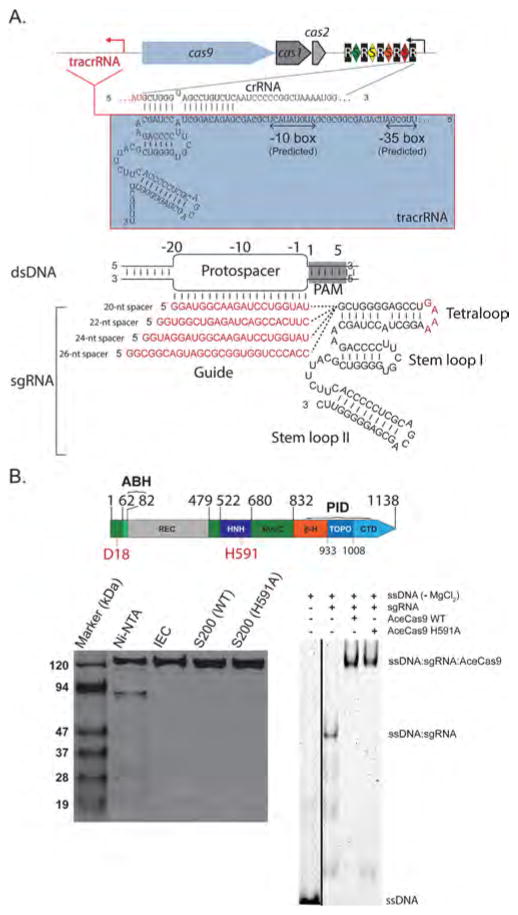
Figure 1.
Identification and purification of the Type II-C Cas9 from A. cellulolyticus (AceCas9). (A) (Top) CRISPR locus in A. cellulolyticus 11B and the sequences of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA). Black rectangles (R), indicate consensus direct repeat sequence and colored diamonds (S) indicate spacer sequences. The red arrow indicates the putative transcription direction for tracrRNA. (Bottom) Schematic representation of R-loop formation between the dsDNA target and the constructed single guide RNA (sgRNA). The guide region of sgRNA (or spacer) is shown in red and base pairs with the targeting DNA strand of the protospacer DNA. Four guide sequences ranging from 20-nts to 26-nts used in this study are listed. (B) (Top) Domain organization for AceCas9. Conserved catalytic residues are indicated in red. RuvC refers to the RuvC nuclease domain (dark green), ABH refers to the Arginine-rich Bridge Helix (light green), REC refers to the heteroduplex Recognition lobe (gray), HNH refers to the HNH nuclease domain (dark blue), PID refers to the PAM-interacting domains and is composed of 3 segments: β-hairpin (β-H, orange), Topo-homology domain (TOPO, blue), and the C-terminal domain (CTD, light blue). (Bottom left) SDS-PAGE analysis of the wild-type (WT) and H591A mutant AceCas9 following Nickel affinity chromatography (Ni-NTA), ion-exchange (IEC) and size-exclusion chromatography (S200). (Bottom right) Binding of AceCas9:sgRNA or H591A:sgRNA complex to single stranded targeting DNA labeled by HEX. Black line indicates the merged lanes from two different gels.