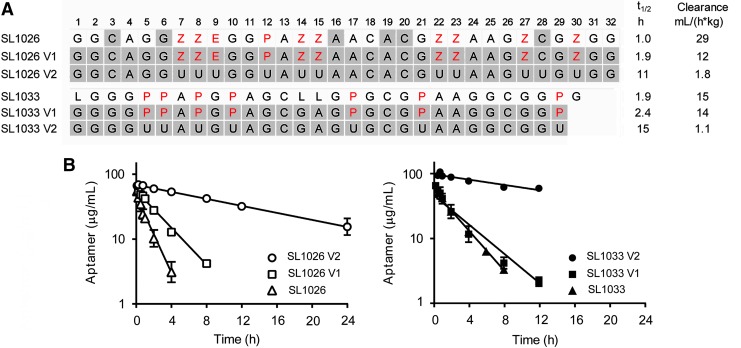
FIG. 1.
Effect of side chains. (A) Sequences (5′→3′) of SL1026, SL1033, and variants thereof, utilized for determination of the effect of side chains on plasma PK. Standard single-letter designations (A, C, G, and U) are used for the unmodified dehydroxy nucleobases, whereas positions designated Z, E, and P show the locations of 5-(N-benzylcarboxamide), 5-[N-(2-phenylethyl)carboxamide], and 5-[N-(1-naphthylmethyl)carboxamide]-modified uridine (highlighted in red text). The positions containing 2′-OMe ribose are shaded gray. All sequences contain a 3′-idT cap and have a 40 kDa PEG attached to the 5′-terminus (not shown). SL1026 contains 2′-OMe ribose at positions 3, 6, 16, 19, 20, and 28, whereas SL1033 contains C3 spacer (L) at positions 1, 14, and 15. The variants (V1 and V2) are fully modified with 2′-OMe ribose. Plasma half-life (t1/2) and plasma clearance values are shown alongside each sequence. (B) Plasma concentration versus time curves. 2′-OMe, 2′-methoxy; PEG, polyethylene glycol; PK, pharmacokinetic.