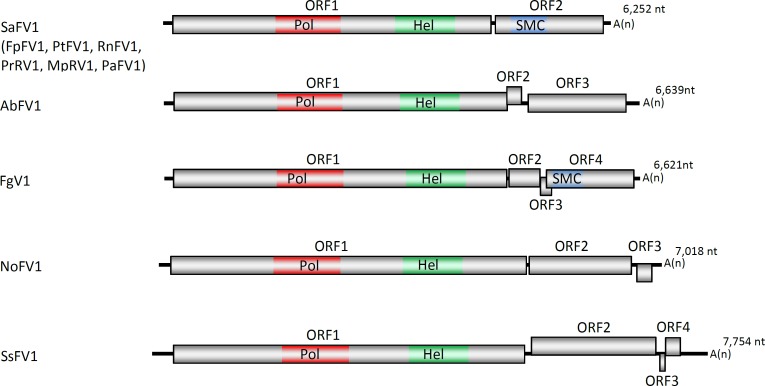
Fig 1. Schematic representation of the putative genomic organization of fusariviruses.
The boxes and lines represent open reading frames (ORFs) and non-coding sequences, respectively. Positions of polymerase (Pol, red), helicase (Hel, green), and structural maintenance of chromosomes (SMC, blue) domains are indicated. Genomes are shown to scale. The A(n) represents poly (A) tail. Sodiomyces alkalinus fusarivirus 1 –SaFV1, Fusarium graminearum virus—FgV1, Sclerotinia sclerotiorum fusarivirus 1—SsFV1, Alternaria brassicicola fusarivirus 1- AbFV1,—Nigrospora oryzae fusarivirus 1—NoFV1. Fusarium poae fusarivirus 1 –FpFV1, Pleospora typhicola fusarivirus 1 –PtFV1, Rosellinia necatrix fusarivirus 1 –RnFV1, Penicillium roqueforti ssRNA mycovirus 1 –PrRV1, Macrophomina phaseolina ssRNA virus 1 –MpRV1, and Penicilium aurantiogriseum fusarivirus 1 –PaFV1, have similar genome organization as SaFV1.