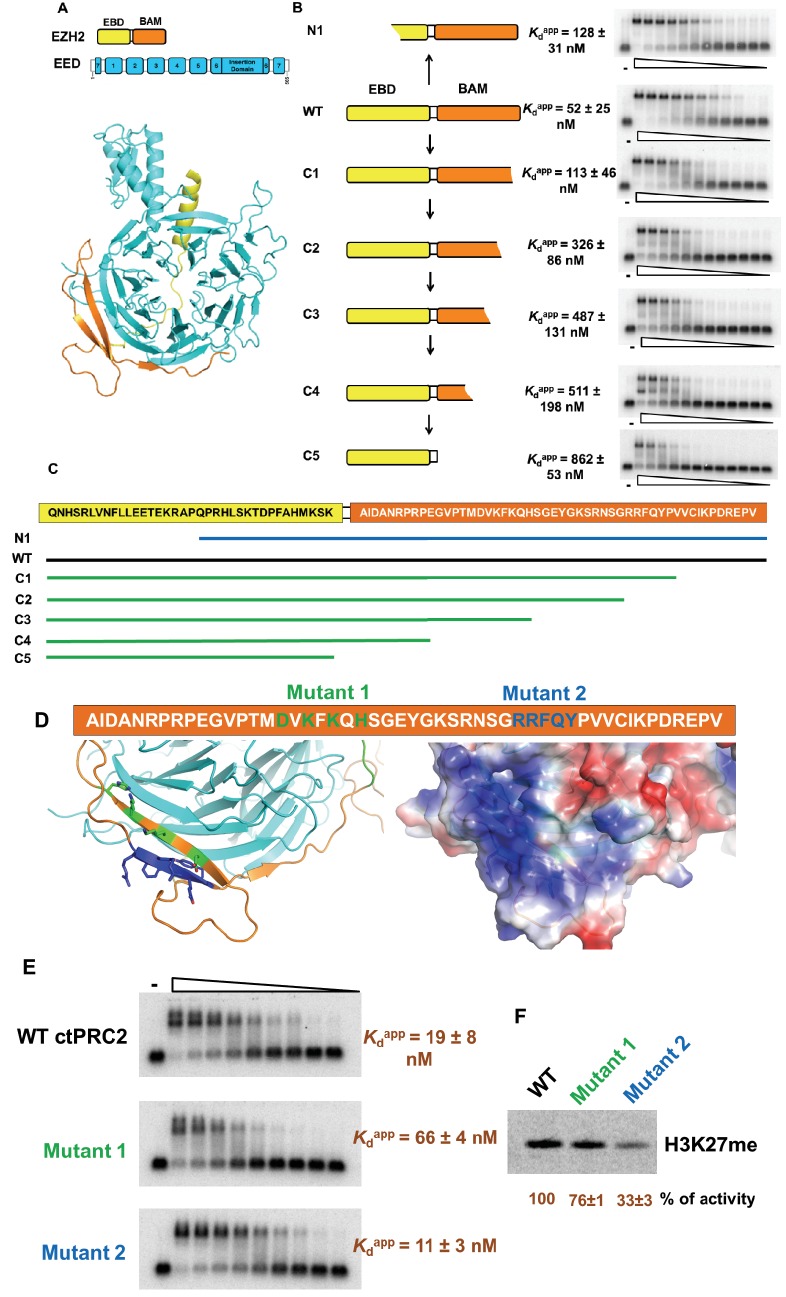
Figure 3. Four residues in BAM domain of ctPRC2 are important for RNA binding.
(A) The minimal RNA-binding complex consists of EED and EBD-BAM domains of EZH2. (B) Mutagenesis study was conducted on the minimal RNA-binding complex, by trimming the EZH2 peptide from the N and C termini. Purified complexes were tested for binding to (GGAA)10 RNA. Threefold titration of proteins was used in each EMSA experiment. Starting concentrations of each complex were: 10 µM for N1, C2, C3 and C4, 5 µM for wild type and C1, and 4 µM for C5. (C) Details of the EZH2 peptide sequence for wild type and the six mutants. Blue line indicates the N-terminal truncation mutant (N1), and green lines refer to the C-terminal truncation mutants (C1-5). (D) Two sets of residues (green and blue) in BAM domain were mutated in the context of the entire ctPRC2. Mutant 1 includes alanine substitution of four residues (shown in green), and mutant 2 includes mutation of five residues (shown in blue) to GGSGG sequence. These residues are also indicated in the structure and the electrostatic map. (E) Wild type and mutant ctPRC2 were tested for binding with (GGAA)10 RNA, and each protein was titrated threefold from 500 nM. (F) Methyltransferase activity on histone H3 was tested for wild type and the two mutants. 14C-labeled SAM and purified H3 were incubated with each protein for 30 min and then resolved on a SDS-PAGE gel. Methyltransferase activity is indicated by the intensity of the radiolabeled H3 band. Activity is normalized to that of wild type, and the mean ±S.D. of three independent experiments is shown.